Abstract
Accurate and precise segmentation of the left atrium (LA) is crucial in the early diagnosis and treatment of atrial fibrillation (AF), which is the most common heart rhythm disease in cases. The size of fibrotic tissue in patients with AF is based on manual examination of images obtained from the gadolinium-enhanced cardiac magnetic resonance imaging (MRI) technique. However, manual examination of the acquired images is time-consuming and has many difficulties, such as LA thickness between observers and resolution according to MR devices. To overcome the challenges of manual segmentation of images obtained from MRI devices, end-to-end, fully automated deep learning-based segmentation architectures have become extremely important today. In this study, an encoder–decoder-based V-shaped deep learning architecture is proposed for precise segmentation of LA. In the proposed architecture, standard convolution and depthwise separable convolution are used together. Thus, sparsely connected blocks with fewer parameters and deeply separable convolutions learn the feature representations better, increasing the robustness of the model. In addition, the bottleneck attention module has been added to each encoder layer, allowing the network to learn which features to focus on and which features to suppress in images by attention mapping channel and spatially. The proposed architecture obtained 0.915 dice and 0.844 Jaccard scores in the STACOM 2018 challenge dataset. The obtained results draw attention to the robustness of the model.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Almost half of all diagnosed cases suffer from atrial fibrillation, the most common cardiac arrhythmia that causes permanent damage to the heart and body [1, 2]. There were 366,000 cardiovascular deaths worldwide in 2021 due to atrial fibrillation and flutter. In addition, by 2021, 8,200,000 cardiovascular deaths have occurred due to atrial fibrillation and flutter [3]. Recurrent atrial attacks cause various problems, such as enlargement of the atrial structure, myofiber changes, and fibrosis [4, 5]. Therefore, it is crucial to precisely segment the atrium in cases with AF so that it can be observed [6]. MRI devices using LGE are one of the best methods to distinguish between scarred and unscarred atrium walls. In addition, LGE MRI is a suitable method for determining the size of fibrotic tissue to examine atrial scar formation after ablation [7,8,9].
Manual segmentation of LA walls obtained from LGE-based MRI is time-consuming and tedious. [8, 10,11,12]. In addition, segmentation results show high variability among experts. Deep learning-based segmentation models have recently become increasingly important to eliminate the manual segmentation difficulties mentioned above and facilitate clinicians’ decision-making [13]. However, many deep learning-based segmentation models add different modules to provide the best performance on data sets, which increases the number of parameters of the model. The large number of parameters of the designed model causes the hardware to be insufficient in terms of calculation. In this study, standard convolution and depthwise separable convolution were used together in the layers to reduce the number of parameters of the deep learning model. As can be seen from experimental studies, the layer-based hybrid structure has significantly reduced the number of parameters of the model. Another situation in medical image segmentation models is that the boundaries of the organs are perceived in the first layers, and the boundary features disappear in the upper layers of the architecture. In organ segmentation, a robust segmentation of the borders of the organ is recommended rather than the segmentation of a sequential structure. The proposed study used the bottleneck attention module (BAM) to prevent the loss of boundary features [14]. The proposed architecture is obtained by applying various additions and subtractions to the V-Net architecture proposed by Milletari et al. [15].
The contributions of the proposed methodology to the literature are as follows:
-
A depthwise separable convolution was added to the second standard convolution layer to solve computational limitations by reducing the number of parameters of the network and learning the feature representations better [16, 17].
-
In the coding layers of the deep learning architecture, the BAM is used, which strengthens the learning performance of the network by emphasizing the high-level features more and suppressing the low-level features after the convolution operations.
-
A fusion loss function increased the network’s test accuracy in LA segmentation from LGE MRI.
-
In addition, as a result of experimental studies, PReLU activation functions were used in the convolution layers and ReLU activation functions in the BAM layer [18, 19].
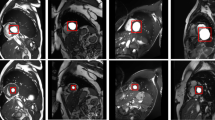
Sample MRI slices of the STACOM 2018 dataset used in training the proposed architecture are shown in Fig. 1. Sample images are pictures and ground truth of the axial images in the data set.
To summarize the remaining sections of the paper, in Sect. 2, brief information is given about the state-of-the-art approaches in the literature on deep learning-based segmentation of atrium images. In Sect. 3, information about the methodology used is given. Section 4 provides information about the data set used for training the model, performance metrics, and parameters used in training. In the fifth section, the experimental analyses of the model are presented and discussed. Finally, in the sixth section, the model's performance is evaluated, and information about future studies is given.
2 Related Works
One of the most critical problems in deep learning-based organ segmentation is that the organ boundaries are considered in the first layers of the deep learning network, and the learning rate of the boundaries decreases in the following layers. In recent studies in the literature, it is seen that this problem is tried to be solved by adding various attention modules, especially in the downsampling stages.
The human visual system performs a series of visual processing to capture important features in any image, focusing the visual system on regions where essential features are [20, 21]. Attention modules have been developed in deep learning architectures to mimic these attention structures of the human visual system and increase segmentation performance. Attention modules focus on high-level features and filter out low-value features [22,23,24,25]. There are many examples of the use of attention modules in deep learning-based segmentation of organs. For instance, Vernikouskaya et al. proposed a U-Net-based fully automatic segmentation model containing a multi-stage pipeline for detecting arrhythmia from cardiac magnetic resonance images (CMRI) [26]. Jabdaragh et al. segmented the left atrium using the multi-task fractal dimension (MTFD-Net) architecture, which combines fractal geometry and a multi-task network. The study tried to increase modeling segmentation performance by mapping images in fractal dimensions [27]. Zhou et al. placed a cross-modal attention module between the encoder and decoder layers for cardiac segmentation, enabling the network to learn interrelated high-level information better [28]. Uslu et al. proposed a multi-task LA-Net network that simultaneously obtains segmentation and edge masks of the left atrium from MRI to detect atrial fibrillation. A combination of cross-attention modules (CAM) and enhanced decoder modules (EDM) were used to incorporate boundary information into the proposed model [29]. Zhang et al. developed three different attention modules in the context of spatial, channel, and region for ventricle segmentation [30]. Zhao et al. segmenting the left atrium focused on tissue boundaries and tissue region. They used attention modules based on the ResNet-101 architecture in the model’s layers. In addition, regional and boundary loss functions are used as hybrids in the model [31]. Li et al. proposed a U-Net architecture with hierarchical aggregation and attention modules for precise segmentation of LA [32].
Additionally, in the literature on left atrium segmentation, Chen et al. proposed a fully automatic segmentation model based on the deep U-Net architecture, which they obtained by modifying the U-Net architecture [33]. Yang et al. used a transfer learning and deep control strategy to focus on spatial dependence in the segmentation region in left atrium images [34]. Uslu and Bharath proposed a segmentation model with a quality control system that uses a single encoder and three decoders and predicts the run-time quality of the segmentation masks [35]. Yang et al. proposed an atlas-based end-to-end segmentation model for segmenting cardiac images obtained from LGE MRI [36]. Tao et al. proposed a fully automated deep learning-based segmentation model for the segmentation of LA and PV [37]. Xiong et al., on the other hand, proposed the AtriaNet architecture, which consists of a multi-scale dual pathway 2-D CNN model for the segmentation of LA [6]. Puybareau et al. have proposed a VGG-Net-based learning transfer model for the segmentation of LA [38].
Many different semi-supervised learning studies in the literature use unlabeled data in medical datasets to train deep learning architectures. One of the recent semi-supervised learning-based studies is the CA-Net architecture proposed by Zhao et al. [39]. In the proposed model, the Trans V module has been added to the V-Net architecture to learn contextual information. Studies in the literature mainly focus on training and testing 2D and 3D deep convolutional neural networks on datasets consisting of LGE MRI images. Luo et al. proposed a deep learning model, which they call U-Net-based semi-supervised uncertainty rectified pyramid consistency (URPC), for the segmentation of medical images [40]. To use abundant unlabeled data in the segmentation of atrium images by Li et al., a module called signed distance map of object surfaces (SDM) was added to the semi-supervised V-Net network used as the backbone [41]. Wang et al. analyzed a semi-supervised learning model called dual-consistency network (DC-Net) on 3D atrium images to achieve high performance in datasets with limited unlabeled data [42]. Luo et al. proposed a new semi-supervised pixel-based dual-task coherence learning strategy (DTCV-Net) that determines the learning strategy from unlabeled data [43]. V-Net architecture constitutes the backbone of the proposed architecture. The major challenge in most of these studies is to segment the LA boundaries and region accurately and robustly. The study suggests a fully automatic pipeline to segment the segmentation region and boundaries more precisely. In addition, the study presents layer-based hybrid convolution as significantly reducing the number of parameters by focusing on computational limitations. The proposed new approach's architectural structure and performance analysis are explained in detail in the remainder of the study.
3 Methodology
The proposed model is an encoder–decoder-based fully automatic V-shaped segmentation architecture that combines standard convolution, depthwise separable convolution, and BAM module.
3.1 Depthwise Separable Convolution
Figure 2 shows the block diagram of the depthwise separable convolution. While the standard convolution performs channel and spatial computations in one step, the depthwise separable convolution consists of two parts: depthwise convolution and pointwise convolution. Depthwise convolution applies a separate convolution to each input channel, while pointwise convolution obtains a linear combination of these convolutions and feeds it to the BAM module. Using depthwise separable convolution instead of standard convolution in the second convolution layer enabled the proposed model to use approximately 20 times fewer parameters. In addition, depthwise separable convolution significantly reduces the computational cost. However, applying depthwise separable convolution in all layers will reduce training accuracy in deep learning architectures where the parameter is very low. The mathematical models clearly show the difference between depthwise separable in Eq. 1 and standard convolution in Eq. 2. In Eqs. 1 and 2, M is the number of input image channels, N is the number of filters, Dp is the image output size, Dk is the number of kernels, and Dg is the size of feature maps for standard convolution.
3.2 Bottleneck attention module (BAM)
The BAM used in the last layers of the downsampling blocks of the proposed architecture is shown in Fig. 3. For the feature map function F ∈ RC×H×W from the depthwise separable convolution layer, BAM infers a 3D attention map M(F) ∈ RC×H×W. Then, the rearranged feature map F' is calculated in Eq. 3.
where ⊗ denotes element-wise multiplication. In the BAM architecture, the attention mechanism and the learning block are now used together to facilitate the gradient flow. First, the channel attention function Mc(F) ∈ RC and spatial attention function Ms(F) ∈ RH×W values are calculated to design an efficient yet powerful module. In addition, σ is a sigmoid function. Then, as seen in Eq. 4, the M(F) attention map was calculated as the sum of the channel and spatial attention values.
3.3 DSBAV-Net
The proposed DSBAV-Net architecture is a V-shaped deep learning network consisting of an encoder and a decoder. The architectural structure of the methodology is shown in Fig. 4. When the proposed model is compared with a baseline V-Net network, after the standard convolution layer with 2 × 2 × 2 filters in each convolutional block, a depthwise separable convolution layer with 3 × 3 × 3 filters was used for depthwise convolution and 1 × 1 × 1 filters were used for pointwise convolution added by removing the second standard convolution layer for feature extraction in depth and spatially to increase the performance of the network. In addition, thanks to the depthwise separable convolution, the network’s parameters have been reduced, and computational limitations have been tried to overcome. In addition, by adding BAM to the last layer of the encoder blocks, the segmentation performance of the network is increased by suppressing the unwanted features and highlighting the high-level features in the feature maps of the images in each layer. A 5 × 5 × 5 convolution is used in the input layer of the proposed model. Only standard convolution and depthwise separable convolution layers were used in the decoder layers. 2 × 2 × 2 filters were used for standard convolution in each block. For depthwise separable convolution, as seen in Fig. 3, 3 × 3 × 3 filters were used for depthwise convolution, and 1 × 1 × 1 filters were used for pointwise convolution. In addition, the ReLU activation function for the BAM performed better, while the PReLU activation function for the other convolutional layers showed higher performance. While creating the layers in the proposed architecture, a fivefold cross-validation method was used.
3.4 Cross-Entropy Dice Fusion Loss Function
A fusion loss, including categorical cross-entropy(CE) and dice loss functions, is proposed to compute the loss of the proposed methodology in this study. Dice loss is 1 − dice score. Dice loss and CE loss functions are shown in Eqs. 6 and 7. While the two loss functions are fused, the loss1 value is obtained by multiplying the dice loss with an α coefficient. The loss2 value was obtained by multiplying the CE loss function with the coefficient 1 − α, and the total cost value was obtained by adding these two loss values. After experimental studies, the ideal α value was determined as 0.5.
True positive (TP) represents correctly predicted lesion pixels, while false positive (FP) represents incorrectly predicted lesion, and false negative (FN) means incorrectly predicted lesion pixels.
In Eq. 8, sp is the score of the positive class, while C gives the number of classes.
4 Materials
4.1 Preparing the Dataset
The segmentation performance of DSBAV-Net has been analyzed in the STACOM 2018 [44] dataset. The STACOM 2018 dataset includes 154 late gadolinium-enhanced (LGE) MRI-based AF images with an isotropic resolution of 0.625 mm × 0.625 mm × 0.625 mm. Of the 154 images, only the ground truth of 100 volumes has been shared publicly. For performance testing of the proposed methodology, the first 60 of the 100 volumes are devoted to training, 20 for testing, and the remaining 20 for validation.
The volumes in the dataset are resized 112 × 112 × 80. The STACOM 18 dataset contains data from different imaging centers. Segmentation masks include the LA region, mitral valve, LA appendage, and parts of the pulmonary vessels. The number of low and high-quality images in this dataset is almost equal.
4.2 Performance Metrics
The segmentation performance of DSBAV-Net was evaluated using the most used performance metrics in the literature, namely dice, intersection over union (IoU), 95% Hausdorff distance (95HD), average surface distance (ASD), precision (Prec) and recall (Rec) [45]. These metrics were calculated with a Python library called MedPy. Equations 5, 11, 12, 13, 14 and 15 give the mathematical equations for the performance criteria. In Eqs. 5, 11, 14 and 15, TP stands for true positive, FP stands for false positive, and FN stands for false negative. Hausdorff distance (HD) measures how far apart two subsets of space are from each other in terms of Euclidean distance. In the formula in Eqs. 12 and 13, A shows the predicted value, and B shows the ground truth, where ‘d(.)’ in the equation is the distance between the points a and b of the predicted set A and the ground truth B. Using 95% HD in the literature eliminates outliers.
4.3 Model’s Training Details
The training details of the model are shown in Table 1. As can be seen in Table 1, the parameters of all networks are optimized with the Adam algorithm and a learning rate of 0.0001 [46]. The training phase was set at 100 iterations for DSBAV-Net and other architectures used for comparative performance analysis. The training activity was manually stopped in case of signs of overfitting. The batch size in the training set was determined as 2 for the STACOM 2018 dataset, and image dimensions were resized to 112 × 112 × 80, respectively. Random crop, center crop, and random rotation (degree 90) and flip (axis = np.random.randint (0, 2)) techniques were applied for the STACOM 2018 dataset. The proposed methodology and all meshes used for comparative analyses were performed on a computer with GTX_1080_TI GPU and Anaconda ecosystem with Pytorch cuda library version 1.13.1.
5 Experimental Analysis and Discussion
5.1 Ablation Study
It has been observed that learning performance decreases when kernel_size is increased in standard convolution layers of DSBAV-Net. Likewise, increasing the kernel size in the depthwise separable convolution layers of DSBAV-Net caused a performance decrease in training with the features of the STACOM 2018 dataset. The imbalance between the quality of the images in STACOM 2018 is one of the most important reasons. The success rate of DSBAV-Net on the STACOM 2018 dataset increased significantly, mainly due to the spatial and channel relevance of the BAM layer. In addition, when PReLU and ReLU activation functions are used in the BAM module, the training and validation losses obtained are shown in Figs. 5 and 6. It was observed that the training performance of DSBAV-Net decreased when the standard convolution layer used in the proposed model was converted into a depthwise separable convolution layer, as shown in Table 2. In addition, the depthwise separable convolution layer used after the standard convolution layer significantly reduced the computational cost.
5.2 Comparative Performance Analysis of the Model
The final training performance results of the proposed methodology and other models are given in Fig. 7. As can be seen from the graph, the proposed architecture consistently increased the training dice score throughout 100 epochs. As can be seen from the training results, DTCV-Net and CA-Net architectures achieved the lowest dice scores.
As can be seen in Table 3, the proposed methodology has been comparatively tested for performance with V-Net, BAM V-Net, depthwise separable V-Net (DSV-Net), CA-Net, URPCU-Net, and DTCV-Net models available in the literature. In a comparative analysis, DSV-Net, one of the architectures, was proposed for the first time in this study. Depthwise separable and standard convolution are used together in each layer of DSV-Net. Hybrid convolution significantly reduced the number of parameters and significantly increased the training and testing speed of the architecture. While choosing the architectures used in the comparative analysis of the proposed methodology, attention was paid to their segmentation performance. Comparative studies, which can be seen in Table 3, show that the proposed model is very robust.
5.2.1 Performance Analysis at STACOM 2018
Table 2 shows the case-based performance results of the proposed architecture in 20 test images on the STACOM 2018 dataset. The proposed architecture showed remarkable success in the HD metric in all cases except cases 2 and 3. The hierarchical attention mechanism of the BAM module of the proposed model resulted in high performance in the HD metric. Successful results were obtained in 20 test images in dice, Jaccard, and ASD metrics.
Table 3 shows comparative performance analyses on the STACOM 2018 data set for the methodology proposed with the latest technology approaches in the literature. As can be seen from the table, the proposed model showed a much better performance than other models. In addition, thanks to layer-based hybrid convolution, the parameters used by the network have been significantly reduced, and the computational limitations caused by the hardware have been tried to be overcome. URPCU-Net, which gave the closest performance to the proposed methodology, achieved high differences such as 1 point in dice score, 2 points in IoU, and 5 points in 95HD. In addition, DSV-Net was first tried in this study, and successful results were obtained.
Figure 8 shows the qualitative analysis between the proposed methodology and the latest technological approaches. The addition of the depthwise separable convolution and BAM to the network ensures that the network is focused on the atrium region. When the proposed model was compared to URPCU-Net, which achieved the closest performance results, the proposed methodology achieved differences of up to 1 point in dice score, 2 points in IoU, and 5 points in 95HD. In addition, DSV-Net was tried for the first time in this study, and successful results were obtained. Additionally, the sections enclosed in the red bounded box in Fig. 8 represent highly inaccurate sections predicted outside the organ region.
Figure 9 also shows the qualitative analysis of the proposed methodology in slices of another test case using state-of-the-art methods. The proposed model performed more robustly than others, especially in section 49 and 50 transitions. Additionally, the sections enclosed in the red bounded box in Fig. 9 represent highly inaccurate sections predicted outside the organ region.
5.3 Discussion
In this section, the advantages and disadvantages of the proposed method are discussed.
5.3.1 Advances in Operation Time
Thanks to the depthwise separable convolution layer in the proposed model, the number of parameters approaching 44.5 million has been reduced to 2.4 million. The proposed model significantly reduces parameters and time, as seen in Table 3. Table 3 shows the step per epoch time in seconds for architectures. One of the reasons for this is that MR images are three-dimensional, and therefore, three-dimensional matrix operations are performed. Additionally, on computers with higher graphics card VRAM size, the operation time can be further reduced by increasing the batch size. In addition, the proposed architecture completed the number of epochs per step almost 30 s earlier than BAMV-Net and baseline V-Net architectures. However, the proposed model fails in the training phase when the standard convolution layer is changed to a depthwise separable convolution layer in the model, as shown.
5.3.2 Segmentation Performance Limitations
Thanks to the BAM module, we observed that the model obtained excellent results even in low-resolution LA images. However, like other models, it had difficulty lowering the HD metric. In addition, the proposed architecture significantly reduces the computational cost compared to the number of parameters of BAM V-Net and baseline V-Net models, as seen in Table 3. On the other hand, when we reduced the number of channels in the architecture, the layer-based hybrid convolution failed to extract high-level features from LA images in the training phase. As can be seen in Fig. 5, the proposed architecture showed superior performance on MRI slices thanks to BAM and depth-separable convolution.
6 Conclusion
This study used standard and depthwise separable convolution for the first time in convolutional blocks. In addition, using the BAM module after each convolutional block in the encoder part of the model, both channel and spatial high-level features are emphasized more, and low-level features are suppressed. In addition, thanks to the depthwise separable convolution, the number of parameters has been reduced approximately 20 times, and both spatial and in-depth monitoring of the features in the image is provided. While the ReLU activation function in the BAM module of the proposed model performed better, the PReLU activation function in the other blocks of the model performed better. In addition, by fusing cross-entropy and dice loss functions, the robustness of the proposed model is increased. A comparative analysis of the proposed model on the STACOM 2018 dataset showed that it is robust. DSBAV-Net achieved a dice score of 91.49 on STACOM 2018 20% test data. The obtained dice score and qualitative analyses also show that the proposed model is highly robust for LA segmentation. The success of the proposed model in other organ segmentations will also be investigated in future studies. In addition, a new loss function that will increase the model’s performance will be discussed.
Data Availability
The [Left Atrium Segmentation] dataset that supports the findings of this study are available in [STACOM 2018 Dataset], [https://drive.google.com/file/d/19uaNSIu3BtXakRECnziG6cnYQVKikwiB/view].
References
Narayan, S.M.; Rodrigo, M.; Kowalewski, C.A.; Shenasa, F.; Meckler, G.L.; Vishwanathan, M.; Baykaner, T.; Zaman, J.A.; Wang, P.: Ablation of focal impulses and rotational sources: What can be learned from differing procedural outcomes? Curr. Cardiovasc. Risk Rep. 11(9), 27 (2017). https://doi.org/10.1007/s12170-017-0552-7
Peng, P.; Lekadir, K.; Gooya, A.; Shao, L.; Petersen, S.E.; Frangi, A.F.: A review of heart chamber segmentation for structural and functional analysis using cardiac magnetic resonance imaging. Magn. Reson. Mater. Phys., Biol. Med. 29(2), 155–195 (2016)
Vaduganathan, M.; Mensah, G.; Turco, J.; Fuster, V.; Roth, G.A.: The global burden of cardiovascular diseases and risk. J. Am. Coll. Cardiol. 80(25), 2361–2371 (2022). https://doi.org/10.1016/j.jacc.2022.11.005
Smaill, B.H.; Zhao, J.; Trew, M.L.: Three-dimensional impulse propagation in myocardium. Circ. Res. 112(5), 834–848 (2013). https://doi.org/10.1161/CIRCRESAHA.111.300157
Xiong, Z.; Fedorov, V.V.; Fu, X.; Cheng, E.; Macleod, R.; Zhao, J.: Fully automatic left atrium segmentation from late gadolinium-enhanced magnetic resonance imaging using a dual fully convolutional neural network. IEEE Trans. Med. Imaging 38(2), 515–524 (2018). https://doi.org/10.1109/TMI.2018.2866845
Malcolme-Lawes, L.C.; Juli, C.; Karim, R.; Bai, W.; Quest, R.; Lim, P.B.; Jamil-Copley, S.; Kojodjojo, P.; Ariff, B.; Davies, D.W.; Rueckert, D.; Francis, D.P.; Hunter, R.; Jones, D.; Boubertakh, R.; Petersen, S.E.; Schilling, R.; Kanagaratnam, P.; Peters, N.S.: Automated analysis of atrial late gadolinium enhancement imaging that correlates with endocardial voltage and clinical outcomes: a 2-center study. Heart Rhythm 10, 1184–1191 (2013)
Marrouche, N.F.; Wilber, D.; Hindricks, G.; Jais, P.; Akoum, N.; Marchlinski, F.; Kholmovski, E.; Burgon, N.; Hu, N.; Mont, L.; Deneke, T.; Duytschaever, M.; Neumann, T.; Mansour, M.; Mahnkopf, C.; Herweg, B.; Daoud, E.; Wissner, E.; Bansmann, P.; Brachmann, J.: Association of atrial tissue fibrosis identified by delayed enhancement MRI and atrial fibrillation catheter ablation: the DECAAF study. JAMA 311, 498–506 (2014)
Zghaib, T.; Nazarian, S.: New insights into the use of cardiac magnetic resonance imaging to guide decision-making in AF management. Can. J. Cardiol. 34, 1461–1470 (2018)
Spragg, D.D.; Khurram, I.; Zimmerman, S.L.; Yarmohammadi, H.; Barcelon, B.; Needleman, M.; Edwards, D.; Marine, J.E.; Calkins, H.; Nazarian, S.: Initial experience with magnetic resonance imaging of atrial scar and co-registration with electroanatomic voltage mapping during atrial fibrillation: success and limitations. Heart Rhythm 9(12), 2003–2009 (2012)
Sohns, C.; Karim, R.; Harrison, J.; Arujuna, A.; Linton, N.; Sennett, R.; Lambert, H.; Leo, G.; Williams, S.; Razavi, R.; Wright, M.; Schaeffter, T.; O’Neill, M.; Rhode, K.: Quantitative magnetic resonance imaging analysis of the relationship between contact force and left atrial scar formation after catheter ablation of atrial fibrillation. J. Cardiovasc. Electrophysiol. 25(13), 138–145 (2014)
Karim, R.; Housden, R.J.; Balasubramaniam, M.; Chen, Z.; Perry, D.; Uddin, A.; Al-Beyatti, Y.; Palkhi, E.; Acheampong, P.; Obom, S.; Hennemuth, A.; Lu, Y.; Bai, W.; Shi, W.; Gao, Y.; Peitgen, H.O.; Radau, P.; Razavi, R.; Tannenbaum, A.; Rueckert, D.; Rhode, K.: Evaluation of current algorithms for segmentation of scar tissue from late gadolinium enhancement cardiovascular magnetic resonance of the left atrium: an open-access grand challenge. J. Cardiovasc. Magn. Reson. 15(1), 105 (2013). https://doi.org/10.1186/1532-429X-15-105
Taghanaki, S. A.; Abhishek, K.; Cohen, J. P.; Cohen-Adad, J.; Hamarneh, G.: Deep semantic segmentation of natural and medical images: a review. arXiv:1910.07655 (2019)
Bernard, O.; Lalande, A.; Zotti, C.; Cervenansky, F.; Yang, X.; Heng, P.A.; Cetin, I.; Lekadir, K.; Camara, O.; Gonzalez Ballester, M.A.; Sanroma, G.; Napel, S.; Petersen, S.; Tziritas, G.; Grinias, E.; Khened, M.; Kollerathu, V.A.; Krishnamurthi, G.; Rohe, M.M.; Pennec, X., et al.: Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Trans. Med. İmaging 37(11), 2514–2525 (2018). https://doi.org/10.1109/TMI.2018.2837502
Park, J.; Woo, S.; Lee, J. Y.; Kweon, I. S.: Bam: Bottleneck Attention Modüle (2018)
Milletari, F.; Navab, N.; Ahmadi, S. A.: V-Net: fully convolutional neural networks for volumetric medical ımage. In: 2016 Fourth International Conference on 3D Vision (2016)
Chollet, F.: Xception: deep learning with depthwise separable convolutions. In: 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp. 1800–1807 (2016)
Hua, B. S.; Tran, M. K.; Yeung, S. K.: Pointwise convolutional neural networks. In: 2018 IEEE/CVF Conference on Computer Vision and Pattern Recognition, Salt Lake City, UT, USA, pp. 984–993 2018. https://doi.org/10.1109/CVPR.2018.00109
He, K.; Zhang, X.; Ren, S.; Sun, J.: Delving deep into rectifiers: surpassing human-level performance on ımagenet classification. In: 2015 IEEE International Conference on Computer Vision (ICCV), pp. 1026–1034
Agarap, A. F.: Deep Learning using Rectified Linear Units (ReLU). arXiv:1803.08375 (2018)
Larochelle, H.; Hinton, G. E.: Learning to combine foveal glimpses with a third-order boltzmann machine. In: Advances in Neural Information Processing Systems, pp. 1243–1251. (2010)
Woo, S.; Park, J.; Lee, J. Y.; So Kweon, I.: Cbam: Convolutional block attention module, pp. 3–19 (2018)
Zhang, P.; Liu, W.; Wang, H.; Lei, Y.; Lu, H.: Deep gated attention networks for large-scale street-level scene segmentation. Pattern Recogn. 88, 702–714 (2019)
Chen, L. C.; Yang, Y.; Wang, J.; Xu, W.; Yuille, A. L.: Attention to scale: scale-aware semantic image segmentation. In: Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, pp. 3640–3649 (2016)
Wang, Y.; Deng, Z.; Hu, X.; Zhu, L.; Yang, X.; Xu, X.; Heng, P. A.; Ni, D.: Deep ttentional features for prostate segmentation in ultrasound. In: Proceeding of IEEE International Conference on Medical Image Computing and Computer Assisted Intervention (MICCAI). Series. LNCS, vol. 11073, pp. 523–530. Springer (2018)
Vernikouskaya, I.; Bertsche, D.; Metze, P.; Schneider, L.M.; Rasche, V.: Multi-network approach for image segmentation in non-contrast enhanced cardiac 3D MRI of arrhythmic patients. Comput. Med. İmaging Graph. 113, 102340 (2024). https://doi.org/10.1016/j.compmedimag.2024.102340
Jabdaragh, A.S.; Firouznia, M.; Faez, K.; Alikhani, F.; Koupaei, J.A.; Gunduz-Demir, C.: MTFD-Net: Left atrium segmentation in CT images through fractal dimension estimation. Pattern Recognit. Lett. 173, 108–114 (2023). https://doi.org/10.1016/j.patrec.2023.08.005
Schlemper, J.; Oktay, O.; Schaap, M.; Heinrich, M.; Kainz, B.; Glocker, B.; Rueckert, D.: Attention gated networks: learning to leverage salient regions in medical images. Med. Image Anal. 53, 197–207 (2019)
Uslu, F.; Varela, M.; Boniface, G.; Mahenthran, T.; Chubb, H.; Bharath, A.A.: LA-Net: a multi-task deep network for the segmentation of the left atrium. IEEE Trans. Med. Imaging 41(2), 456–464 (2022). https://doi.org/10.1109/TMI.2021.3117495
Zhou, Z.; Guo, X.; Yang, W.; Shi, Y.; Zhou, L.; Wang, L.; Yang, M.: Cross-modal attention-guided convolutional network for multi-modal cardiac segmentation. In: Proceedings of the International Workshop on Machine Learning in Medical Imaging, Series on LNCS, vol. 11861, pp. 601–610. Springer (2019)
Zhang, T.; Li, A.; Wang, M.; Wu, X.; Qiu, B.: Multiple attention fully convolutional network for automated ventricle segmentation in cardiac magnetic resonance imaging. J. Med. Imaging Health Inform. 9(5), 1037–1045 (2019). https://doi.org/10.1166/jmihi.2019.2685
Zhao, Z.; Puybareau, É.; Boutry, N.; Géraud, T.: Do not treat boundaries and regions differently: an example on heart left atrial segmentation. In: 2020 25th International Conference on Pattern Recognition (ICPR), Milan, Italy, pp. 7447–7453 (2021). https://doi.org/10.1109/ICPR48806.2021.9412755
Li, C.; Tong, Q.; Liao, X.; Si, W.; Sun, Y.; Wang, Q.; Heng, P. A.: Attention-based hierarchical aggregation network for 3D left atrial segmentation. In: International Workshop Statistical Atlases and Computational Models of the Heart, pp. 255–264. Springer (2018). https://doi.org/10.1007/978-3-030-12029-0_28
Chen, C.; Bai, W.; Rueckert, D.: Multi-task learning for left atrial segmentation on GE-MRI. In: Pop, M. et al. Statistical Atlases and Computational Models of the Heart. Atrial Segmentation and LV Quantification Challenges. STACOM 2018. Lecture Notes in Computer Science (), vol. 11395. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-12029-0_32
Yang, X.; Wang, N.; Wang, Y.; Wang, X.; Nezafat, R.; Ni, D.; Heng, P.: Combating Uncertainty with Novel Losses for Automatic Left Atrium Segmentation. In: Pop, M., et al. Statistical Atlases and Computational Models of the Heart. Atrial Segmentation and LV Quantification Challenges. STACOM 2018. Lecture Notes in Computer Science(), vol. 11395. Springer, Cham (2019). https://doi.org/10.1007/978-3-030-12029-0_27
Uslu, F.; Bharath, A.A.: TMS-Net: a segmentation network coupled with a run-time quality control method for robust cardiac image segmentation. Comput. Biol. Med. 152, 106422 (2023). https://doi.org/10.1016/j.compbiomed.2022.106422
Yang, G.; Zhuang, X.; Khan, H.; Haldar, S.; Nyktari, E.; Li, L.; Wage, R.; Ye, X.; Slabaugh, G.; Mohiaddin, R.; Wong, T.; Keegan, J.; Firmin, D.: Fully automatic segmentation and objective assessment of atrial scars for long-standing persistent atrial fibrillation patients using late gadolinium-enhanced MRI. Med. Phys. 45(4), 1562–1576 (2018). https://doi.org/10.1002/mp.12832
Tao, Q.; Ipek, E.G.; Shahzad, R.; Berendsen, F.F.; Nazarian, S.; van der Geest, R.J.: Fully automatic segmentation of left atrium and pulmonary veins in late gadolinium-enhanced MRI: towards objective atrial scar assessment. J. Magn. Reson. Imaging 44, 346–354 (2016)
Puybareau, É.; Zhao, Z.; Khoudli, Y.; Carlinet, E.; Xu, Y.; Lacotte, J., : Left atrial segmentation in a few seconds using fully convolutional network and transfer learning. In: International Workshop Statistical Atlases and Computational Models of the Heart, pp. 339–347. Springer (2018). https://doi.org/10.1007/978-3-030-12029-0_37
Zhao, C.; Xiang, S.; Cai, Z.; Shen, J.; Li, S.; Zhou, S.; Zhao, D.; Su, W.; Guo, S.; Wang, Y.: Context-aware network for semi-supervised segmentation of 3d left atrium (2023). https://doi.org/10.2139/ssrn.4087641
Luo, X.; Wang, G.; Liao, W.; Chen, J.; Song, T.; Chen, Y.: Semi-supervised medical image segmentation via uncertainty rectified pyramid consistency. Med. Image Anal. 80, 102517 (2022). https://doi.org/10.1016/j.media.2022.102517
Li, S.; Zhang, C.; He, X.: Shape-aware semi-supervised 3D semantic segmentation for medical ımages. In: Martel, A. L., et al. Medical Image Computing and Computer Assisted Intervention – MICCAI 2020. MICCAI 2020. Lecture Notes in Computer Science(), vol 12261. Springer, Cham (2020). https://doi.org/10.1007/978-3-030-59710-8_54
Wang, J.; Liu, X.; Yin, J.; Ding, P.: DC-net: dual-consistency semi-supervised learning for 3D left atrium segmentation from MRI. Biomed. Signal Process. Control 78, 103870 (2022). https://doi.org/10.1016/j.bspc.2022.103870
Luo, X.; Chen, J.; Song, T.; Wang, G.: Semi-supervised medical image segmentation through dual-task consistency. In: Proceeding of the AAAI Conference on Artificial İntelligence (AAAI), vol. 35, no. 10, pp. 8801–8809. Palo Alto, CA, USA (2021)
Xiong, Z.; Xia, Q.; Hu, Z.; Huang, N.; Bian, C.; Zheng, Y.; Vesal, S.; Ravikumar, N.; Maier, A.; Yang, X.; Heng, P.A.; Ni, D.; Li, C.; Tong, Q.; Si, W.; Puybareau, E.; Khoudli, Y.; Géraud, T.; Chen, C.; Bai, W., et al.: A global benchmark of algorithms for segmenting the left atrium from late gadolinium-enhanced cardiac magnetic resonance imaging. Med. İmage Anal. 67, 101832 (2021). https://doi.org/10.1016/j.media.2020.101832
Kasmaiee, S.; Homayounpour, M.: Correcting spelling mistakes in Persian texts with rules and deep learning methods. Sci. Rep. 13, 19945 (2023). https://doi.org/10.1038/s41598-023-47295-2
Ba, J.; Kingma, P.: Adam: a method for stochastic optimization. In: International Conference on Learning Representations (ICLR), pp.1–11 (2015)
Funding
Open access funding provided by the Scientific and Technological Research Council of Türkiye (TÜBİTAK).
Author information
Authors and Affiliations
Contributions
Hakan Ocal contributed to research, conceptualization, preparing the dataset, validation, determining methodology, writing code, visualizing data, drafting the paper, writing the paper, reviewing, and editing. There is no other organization or person contributing to this paper.
Corresponding author
Ethics declarations
Competing interests
The author declares that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Ocal, H. DSBAV-Net: Depthwise Separable Bottleneck Attention V-Shaped Network with Hybrid Convolution for Left Atrium Segmentation. Arab J Sci Eng (2024). https://doi.org/10.1007/s13369-024-09131-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13369-024-09131-1