Abstract
We used DNA metabarcoding to assess the seasonal diets of the large Japanese wood mouse, Apodemus speciosus (Muridae, Rodentia), in forest edges adjacent to citrus orchards on Innoshima Island, Japan. We used one chloroplast and three mitochondrial DNA barcoding markers to determine mouse diets. Among the various plant and invertebrate diets, A. speciosus typically consumed Chinese cork oak (Quercus variabilis) in early spring (likely acorns preserved during winter) and gypsy moths (Lymantria dispar, a forest pest) in late spring and summer. In addition, we found that A. speciosus also preyed on orchard pests, including the gutta stink bug and other potentially harmful invertebrates. The season during which A. speciosus preyed on stink bugs corresponded with the harvest of orchard products. This study revealed several of the ecological roles of A. speciosus within the boundary zone between forest and human ecosystems. Furthermore, based on the performance of various mitochondrial markers in dietary profiling of invertebrate food items, we recommend the multi-locus DNA metabarcoding method to comprehensively assess the diet of A. speciosus.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Assessing the ecological roles of organisms in forest ecosystems is crucial for understanding how forest-dwelling species promote forest sustainability and provide essential ecosystem services for humans (Brockerhoff et al. 2017). Such endeavors are especially important given accelerating global deforestation (Hansen et al. 2013) as well as the urgency of meeting the United Nations Sustainable Development Goals over the next decade, particularly target 15 (Life on Land; https://sdgs.un.org/goals/goal15, accessed 1 June 2021). In Japan, two-thirds of the land base is forested. These forests are fundamental to the country’s biodiversity (Hill et al. 2019) but are unique in experiencing the extremely high-intensity anthropogenic pressures exerted over a small land area. Therefore, identifying the ecological roles of forest-dwelling species is fundamental for building a society that exists in harmony with nature and preserving forest sustainability.
The large Japanese wood mouse, Apodemus speciosus (Muridae, Rodentia), is endemic to Japan. This species is one of the most ancient mammals in Japan (Sato 2016) and is widely distributed in forests as well as at the boundary between forests and nearby human settlements from northern Hokkaido to southern Honshu, Shikoku, Kyushu, and other adjacent small islands (Ohdachi et al. 2015). Previous studies have documented conflicts due to the interactions between human infrastructure (roads, buildings, or city urbanization) and the forest ecosystem based on wood mouse genetics (Hirota et al. 2004; Sato et al. 2014, 2020). Thus, A. speciosus represents a wildlife species that can be studied to elucidate specific aspects of the interplay between forest and anthropogenic ecosystems.
One strategy for assessing interrelationships between forest and anthropogenic ecosystems is food-web analysis involving the assessment of wildlife diets. Previous morphological analyses of A. speciosus feces and stomach contents were unable to adequately identify fragments of prey species, even with the aid of a microscope (Abe and Oya 1974; Mizushima and Yamada 1974; Tatsukawa and Murakami 1976; Tachibana et al. 1988). More recently, a DNA metabarcoding approach, based on high-throughput sequencing, was used to determine the diets of wood mice (A. speciosus and/or A. argenteus) on Hokkaido and islands in the Seto Inland Sea; this allowed for dietary comparisons under interspecific competition and across geographic space (Sato et al. 2018, 2019). Sato et al. (2019) assessed A. speciosus diets on several islands in the Seto Inland Sea, but this study was temporally limited and did not characterize seasonal variation. Documenting seasonal variation in diets would provide more insight into the ecological role of A. speciosus within forest edges adjacent to human settlements.
Applications of DNA metabarcoding in diet studies have been increasing, and several promises and pitfalls are associated with this approach (de Sousa et al. 2019; Ando et al. 2020). A key issue is the lack of consensus concerning the choice of primers to use in analyses. The mitochondrial cytochrome c oxidase subunit I (COI) and 16S ribosomal RNA (16S) genes are the most frequently targeted genomic regions for DNA metabarcoding of invertebrates. Accordingly, several universal primers have been developed for these regions (Tournayre et al. 2020). Despite their name, universal primers are not always strictly universal due to differences in the polymerase chain reaction (PCR) efficiency of each primer based on the target species. This issue leads to variation in the detection of dietary components among primers (Tournayre et al. 2020; Browett et al. 2021). To date, researchers do not agree on whether to use multiple primers for a single locus or to combine primers for multiple loci (Alberdi et al. 2018; Corse et al. 2019; Browett et al. 2021) or to use a single, well-designed primer (Elbrecht et al., 2019; Tournayre et al. 2020). Therefore, the performance of each barcoding primer should be assessed to judge which is appropriate for each case study. However, such a comparison of marker performances has not yet been conducted for DNA metabarcoding analyses involving rodents. In fact, no studies have examined how primer choice affects the representation of seasonality for any organism.
In this study, we used a DNA metabarcoding approach for fecal samples by employing several short barcoding markers for the target regions (shorter markers are better for degraded fecal samples; Vamos et al. 2017; Tournayre et al. 2020). Our objectives were to identify seasonal variation in the plant and invertebrate diet of A. speciosus on Innoshima Island, Japan, and to elucidate its roles in forest and agro-ecosystems. We also discuss primer selection based on comparisons of three mitochondrial DNA primers for invertebrates.
Materials and methods
Study site and obtained samples
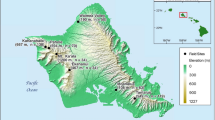
Innoshima Island was not included in previous studies of the A. speciosus diet on islands in the Seto Inland Sea (Sato et al. 2019). The present study took place at the base of Mt. Okuyama, near the town of Mukunoura, Innoshima Island, Hiroshima Prefecture, Japan (Fig. 1). The island is located within a typical temperate zone climate. Approximately one-third of the island area (1251.96 of 3976 ha) is dominated by forest, and 40% of the forested area is composed of broad-leaved trees. The forest near Mukunoura is dominated by deciduous tree species, with high prevalence of Chinese cork oak (Quercus variabilis). Citrus orchards, typically Citrus hassaku, occur near the forest study site in Mukunoura (Fig. 1; citrus fruit agriculture is a major industry on the island). This island therefore provides a suitable location for assessing the interplay between forest and agro-ecosystems.
We captured 19 individuals of A. speciosus using 40–60 Sherman live traps set in a 50 × 50-m area from March to August 2019. We sampled wood mice every month to assess seasonal differences in diet. The trapping period included the rainy season (called Tsuyu in Japan; Table 1, late June to late July). At the study site, the Tsuyu season occurred from 26 June to 25 July in 2019. The capture rate of wood mice declines dramatically in September of every year in our study area (personal observation of the first author); therefore, we stopped sampling in September. Traps baited with oats (Avena sativa) were set out for one night. Upon capture, we collected several tissue samples using an ear punch for genetic species identification (see below) and then released the individuals. Each mouse sample was collected from different individuals, because ear-punched individuals were never re-sampled. We transported traps to the laboratory within 30 min of release and collected feces in the trap using sterilized tweezers. Feces were collected into 1.5-ml plastic tubes. Fecal samples were then preserved at − 80 °C until DNA extraction. We obtained permission from Hiroshima Prefecture to collect the wood mice samples in the Innoshima island (no approval code from the prefecture) and conducted protocols approved by the Animal Care and Use Committee of Fukuyama University (approval code: H30-Animal-8). We followed the guidelines of the Procedure of Obtaining Mammal Specimens established by the Mammal Society of Japan (https://www.mammalogy.jp/en/guideline.pdf; accessed 1 June 2021).
Avoidance of contamination
For all molecular experiments described below, we used filtered tips and experimental globes. Before performing each experiment, we wiped the laboratory bench with DNA Away (Thermo Scientific, Waltham, MA, USA) to remove any remaining DNA. DNA extraction and PCR for both tissue and fecal samples were performed in the same laboratory (but at different benches), and sequencing occurred in a different building. We conducted negative control experiments for PCR and sequencing as described below to control for contamination.
DNA extraction
We used two commercial DNA extraction kits, the DNeasy Blood & Tissue Kit and QIAamp DNA Stool Mini Kit (Qiagen, Hilden, Germany), to extract DNA from ear tissue and fecal samples, respectively, following the manufacturer’s instructions. Prior to extraction, three pieces of feces were chopped as finely as possible using sterilized surgical scissors within each tube. Following DNA extraction from both tissue and fecal samples, the sample concentration of DNA was calculated using a NanoDrop (Thermo Fisher Scientific, Waltham, MA, USA) and a Qubit fluorometer (Thermo Fisher Scientific), respectively, to adjust the DNA concentration to 100 ng/µL and 5 ng/µL in tissue and fecal samples, respectively, for PCR.
Species identification
We followed methods in Sato et al. (2014) to determine a partial nucleotide sequence of the Dloop region in the mitochondrial DNA using the Sanger DNA sequencing method for the collected samples. We then compared the obtained sequences with previously determined sequences through a BLASTn search (Altschul et al. 1990) for species identification.
DNA metabarcoding
We performed two-step PCRs to prepare libraries for subsequent next-generation sequencing (NGS) analyses. All PCR amplifications described below were performed using an automated thermal cycler (LifeTouch thermal cycler; Bioer Technology, Hangzhou, China). In each PCR, we targeted three molecular markers in the mitochondrial DNA, one of COI and two of 16S (16S-epp and 16S-kp), for invertebrate dietary profiling, and one molecular marker in the chloroplast DNA, the P6 loop of the trnL (UAA) intron (trnL), for plant dietary profiling (Table 2). We used these primers based on their performance in previous studies on wood mice (Sato et al. 2018, 2019) and published reviews (Alberdi et al. 2017; Ando et al. 2020). In the first PCR, we amplified each target region using KAPA HiFi Hotstart Ready Mix (Kapa Biosystems Inc., Wilmington, MA, USA). The first PCR reaction mixture (25 μL) contained 2 × KAPA HiFi HotStart ReadyMix, 0.3 μM of each universal primer (Table 2), and template DNA (20–45 ng), and was adjusted using PCR-grade water as necessary. We used specific primers targeted to the COI, 16S-epp, 16S-kp, and trnL genomic regions (Table 2). The 3′-end of each primer used in the first PCR had an adapter sequence targeted by the second PCR and sequencing primers. Between the specific and adapter primer sequences, there were six base-pair (bp) sequences (6 N) for efficient sequencing on the Illumina MiSeq NGS platform (Illumina, San Diego, CA, USA). The sequences of the first PCR primers were 5′-ACACTCTTTCCCTACACGACGCTCTTCCGATCTNNNNNN***-3′ (*** represents the first PCR forward primers; Table 2) and 5′- GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCTNNNNNN + + + -3′ (+ + + represents the first PCR reverse primers; Table 2). Therefore, the total target length included the length of each specific primer, a 79-bp adapter, and 6-bp sequences (Table 2).
The first PCR conditions for COI and 16S-epp were as follows: initial denaturation at 95 °C for 15 min, followed by 35 cycles of denaturation at 98 °C for 20 s, annealing at 57 °C for 90 s, and extension at 72 °C for 60 s, and a final extension at 72 °C for 10 min. The first PCR conditions for 16S-kp were as follows: initial denaturation at 95 °C for 15 min, followed by 40 cycles of denaturation at 98 °C for 20 s, annealing at 50 °C for 60 s, and extension at 72 °C for 30 s, and a final extension at 72 °C for 5 min. Finally, the first PCR conditions for trnL were as follows: initial denaturation at 95 °C for 15 min, followed by 40 cycles of denaturation at 98 °C for 20 s, annealing at 57 °C for 90 s, and extension at 72 °C for 60 s, and a final extension at 72 °C for 5 min. We confirmed that there was no amplification of the negative control samples (one per 4–8 samples) using agarose gel electrophoresis analyses. The products of the first PCR were purified using AMpure XP beads (Beckman Coulter Inc., Brea, CA, USA).
In the second PCR, we amplified each target region using KAPA HiFi Hotstart Ready Mix and a pair of primers that contained adapters (P5 and P7) and indices as sample identifiers. The PCR mixture (24 μL) contained 2 × KAPA HiFi HotStart ReadyMix, 0.29 μM of each index primer below, and 2 μL of a purified first PCR product, and was adjusted using PCR-grade water as necessary. The primers for the second PCR were designed to be attached to the 5′ end of the first PCR product, and they contain the index sequence in the middle part and the P5 or P7 adapter at the 5′ end: [forward primer] 5′-AATGATACGGCGACCACCGAGATCTACAC-[8-bp index]-ACACTCTTTCCCTACACGACGCTCTTCCGATCT-3′ and [reverse primer] 5′-CAAGCAGAAGACGGCATACGAGAT-[8-bp index]-GTGACTGGAGTTCAGACGTGTGCTCTTCCGATCT-3′. A 69-bp sequence was therefore added to the first PCR products after the second PCR. The conditions of the second PCR were as follows: initial denaturation at 95 °C for 3 min, followed by 12 cycles of denaturation at 98 °C for 20 s, annealing at 55 °C for 15 s, and extension at 72 °C for 15 s, and a final extension at 72 °C for 5 min. We confirmed that there was no amplification of the negative control samples (one per 4–8 samples) using agarose gel electrophoresis. The second PCR products were mixed on an equi-volume basis, wherein negative control samples were also included, and purified using AMpure XP beads.
Following purification, gel electrophoresis was performed using E-Gel™ SizeSelect™ II Agarose Gels 2% to extract target DNA fragments (Thermo Fisher Scientific). We then assessed the concentration and quality of the extracted samples using a Qubit fluorometer and an Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA, USA), respectively. Finally, we conducted several runs of sequencing using Illumina MiSeq with Nano or Micro 300-cycle reagent kits with a 30% PhiX control spike-in in each run.
Data filtering and homology search
We used Claident (Tanabe and Toju 2013) for data filtering and conducted a DNA database search using BLASTn for dietary profiling. The analysis codes for data processing are provided in Supplementary Information SI1. First, the bcl file obtained from MiSeq was converted to a FASTQ file using the bcl2fastq command. For the FASTQ file, DNA sequence data were demultiplexed based on an 8-bp index and the primer regions and their flanking sequences were removed using clsplitseq. Subsequently, forward and reverse paired-end sequences were merged using clconcatpair. Low-quality data, noisy data, too-short sequences, and chimeras were filtered using clfilterseq and clcleanseqv. Filtered sequences were clustered using clclassseqv with a 98% minimum identity to obtain operational taxonomic units (OTUs). Sequences that were noisy but found to be similar to an OTU cluster were recovered using clrecoverseqv. Chimeras were searched for again using a specific database and were removed using clrunuchime (except for trnL). For these filtered OTU sequences, a BLASTn search for dietary OTUs in the DNA database (the animal COI and animal mtDNA databases for COI and 16S, respectively, as available in Claident by default and in the cpDNA database for trnL constructed in this study through a search with the term “chloroplast” in the NCBI nucleotide database) was performed using clidentseq, and finally, lowest common ancestor (LCA) analyses were performed using classigntax. OTUs that had fewer than 10 reads were classified as low-frequency OTUs and removed (Sato et al. 2018, 2019). When we could not identify a certain level of taxonomic resolution in the LCA analyses, we repeated the BLAST search individually to confirm top hit sequences. Raw data, in the form of FASTQ files that had been subjected to the clsplitseq analyses, are available from DRA (DDBJ Sequence Read Archive) under the accession number DRA011398.
Rarefaction and statistical analyses
Although there was no evidence of amplification in the negative controls, some minor reads were assigned to them. Therefore, the number of reads detected in the negative controls was removed from those of other samples. For example, if a sample and a negative control had 500 and 10 reads for OTU1, respectively, we considered the read number obtained from the sample to be 490 (500 − 10). We also removed infrequent reads, i.e., those accounting for < 0.1% of the total number of reads per sample. We then used rarefaction to account for differences in the number of reads obtained per sample (Table 1) in each marker individually with the package vegan (Oksanen et al. 2019) in R (R Core Team 2020). We determined saturation on the rarefaction curves at a selected number of reads, where the selected number was based on either the minimum (3900, 10,000, and 20,000 for COI, 16S-epp, and 16S-kp, respectively) or a smaller arbitrary (6000 for trnL) number. We confirmed that the rarefaction curves were saturated at the points of the pre-determined threshold values reported above (Supplementary Information SI2). We calculated pairwise Bray–Curtis and Jaccard indices, which represent differences in diet between sampled individuals, based on relative read abundance data and binary (0/1) occurrence data, respectively. The obtained matrices were used to construct a non-metric multidimensional scaling (NMDS) plot, again using the package vegan in R. We divided samples into two seasons, spring (March, April, May, and June) and summer (July and August), based on the timing of the rainy (Tsuyu) season, and assessed differences in diet between seasons using PerMANOVAs with 10,000 permutations.
Results
Species identification
We confirmed that all sequences of the mitochondrial Dloop region from each of the 19 individuals matched those of A. speciosus using a BLASTn search. For samples 6, 7, 10, 15, 19, and 21, a novel haplotype (Inn4) was obtained. For all other samples, the haplotypes matched those from previous findings (Inn1 and Inn2; Sato et al. 2017). The obtained sequences were deposited in the DDBJ/ENA/GenBank international DNA database under accession numbers LC600288–LC600306 (Table 1).
Dietary profiling
We obtained read data for the COI, 16S-epp, 16S-kp, and trnL barcoding markers from 19, 19, 15, and 16 wood mice, respectively, after next-generation sequencing analyses (Table 1). For invertebrates, rarefied COI, 16S-epp, and 16S-kp data indicated the presence of 230, 20, and 21 OTUs, respectively. Among these, 16S-epp and 16S-kp data included host reads, in which 98% and 94% of the total number of reads, respectively, were indicative of Murinae species and were assumed to belong to the host (A. speciosus). We then excluded these host reads from further analyses. We also removed reads for a nematode from both COI and 16S-epp data, on the assumption that nematodes are not a component of wood mouse diets, as they are sometimes observed on the skin of the wood mouse (personal observation of the first author). Finally, we removed samples 3, 8, 9, 15, 16, 17, 20, and 21 from 16S-epp analyses and sample 15 from 16S-kp analyses, as the number of reads became zero for all OTUs after the rarefaction analyses. The OTUs were reduced to zero because the relative read abundance for the host wood mouse (removed later) was predominant for these 16S markers, and the reads for prey taxa were not sampled in the rarefaction analyses. For plants, we first removed samples 9, 11, 12, and 15 from rarefaction analyses of trnL due to a low number of reads relative to other samples (< 6000 reads/sample; Table 1). The remaining rarefied trnL data included 63 OTUs. Among these, 38.8% of the total rarefied reads were assigned to Avena sp., which was assumed to be the bait used in this study, A. sativa, and removed from further analyses.
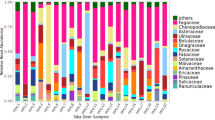
The NMDS plot of COI rarefied relative read abundance data (stress value [sv] = 0.13) indicated a seasonal difference between spring and summer (Fig. 2A), which was supported by the results of the PerMANOVA (P < 0.01, F = 3.41, R2 = 0.17). The occurrence data also supported the presence of a seasonal difference in the NMDS plot (Fig. 2B; sv = 0.15) and PerMANOVA (P < 0.01, F = 2.17, R2 = 0.11). The majority of the rarefied reads (77.9%) were assigned to Lepidoptera (Supplementary Information SI3). The major lepidopteran families included Erebidae (6.4% of the Lepidoptera reads), Lymantriidae (7.7%), and Noctuidae (5.9%) (Fig. 3A). Although we used the family name Lymantriidae based on the registered name in the DDBJ/ENA/Genbank DNA database, we note that this family is currently considered a subfamily (Lymantriinae) in Erebidae (Zahiri et al., 2010). Moths that could be identified at the species level and also occur in Japan included Pidorus glaucopis (Zygaenidae); Trachea atriplicis, Mythimna separata, and Melanchra persicariae (Noctuidae); and Lymantria monacha and L. dispar (Lymantriidae). In particular, L. dispar, the gypsy moth, was commonly observed in samples from June and July, representing 78.0% of the total reads for the family (Supplementary Information SI4). Although we did not achieve species-level identification for Noctuidae, we note that the genus Lithophane accounted for 74.9% of the total reads for the family. We were unable to identify genera or species belonging to Erebidae, but this family was a major component of moth prey in spring (Fig. 3). The second most frequently encountered OTU, at the order level, was Diptera, accounting for 18.1% of the total number of reads. Of these, Chironomidae accounted for 65.0% and Tachinidae accounted for 25.4% (Supplementary Information SI3). Nearly all of the reads belonging to Chironomidae were from the species Tanytarsus formosanus (98.4%), but we were unable to obtain genus- or species-level identification for Tachinidae. Finally, beetles (Carabidae) were most frequently detected in July (Fig. 3; Supplementary Information SI4), for which the majority of reads belonged to Carabus (72.2%). We also detected a species of Vespula (Vespidae), but only during the summer season (July and August; Supplementary Information SI4).
Non-metric multidimensional scaling (NMDS) plot based on Bray–Curtis (A, C, E) and Jaccard (B, D, F) similarity indices for rarefied read data obtained from COI (A, B), 16S-epp (C, D), and 16S-kp (E, F) analyses. Open circles and open diamonds indicate samples collected in spring and summer, respectively
Read numbers assigned to each taxonomic category (orders and families) for COI (A), 16S-epp (B), and 16S-kp (C). Before plotting, the read numbers were rarefied and therefore normalized among samples to a pre-determined value for each marker as explained in the text. Chironomidae (Diptera) and unassigned Lepidoptera with very large read numbers (7778 primarily in August and 40,057 distributed throughout the sampling period, respectively) for COI were removed to show the other read data clearly; see Supplementary Information SI3 for those additional data. Reads associated with an unassigned order, Murinae (presumably A. speciosus), and a nematode were excluded. We performed rarefaction on spring observations to account for differences in sample sizes between spring and summer
For the 16S-epp rarefied relative read abundance data, the NMDS plot, based on Bray–Curtis distances, did not indicate seasonal differences in diet (Fig. 2C; sv = 0.13), nor did the results of the PerMANOVA (P > 0.05, F = 0.83, R2 = 0.084). Similarly, the occurrence data did not indicate a seasonal difference in either the NMDS plot (Fig. 2D; sv = 0) or PerMANOVA (P > 0.05, F = 1.01, R2 = 0.08). Broadly, from 16S-epp data, dietary components different from those from COI were detected, specifically Hemiptera (stink bugs) and Orthoptera (grasshoppers) (Fig. 3B). Within Hemiptera, we detected a Kempfer cicada (Platypleura sp.), which we assumed to be Platypleura kaempferi, given that only one species of this genus occurs in the study area, and the gutta stink bug Physopelta gutta (Supplementary Information SI5). Within Orthoptera, we detected a bush-cricket, Tettigonia sp. (Supplementary Information SI5). In contrast to the COI reads, Geometridae was the most frequently encountered moth family according to the rarefied 16S-epp data (Fig. 3B). Among the Geometridae OTUs, we detected the genus Biston (Supplementary Information SI5). A re-analysis of the BLASTn search uncovered identifications for a Biston sp., Inurois fletcheri, and Wilemania nitobei, none of which were assigned in the LCA analysis. The most frequently detected Geometridae OTU was not identified at the genus level. Similar to the results using COI, Lymantriidae (Lymantria) was observed in June (Supplementary Information SI5).
The NMDS plot using 16S-kp rarefied data indicated that samples 13 and 14 were outliers for both relative read abundance and occurrence data (Supplementary Information SI6). These two samples did not share any OTUs with the other samples (only Lymantria dispar and two unassigned Lepidoptera taxa in samples 13 and 14, respectively, were detected; Supplementary Information SI7); therefore, these two samples were removed from the analyses. After removing these samples, the NMDS plot based on the relative read abundance data (sv = 0.0001) suggested a seasonal difference in diet (Fig. 2E), which was confirmed using a PerMANOVA (P < 0.05, F = 2.0, R2 = 0.17). The occurrence data supported the presence of a seasonal difference in the NMDS plot (Fig. 2F; sv = 0.06), although the PerMANOVA outcome was not significant (P > 0.05, F = 1.20, R2 = 0.11). Dietary components in the spring samples were dominated by unassigned Lepidoptera (Fig. 3C). However, the summer samples were dominated by Cicadidae (Hemiptera; Fig. 3C; Supplementary Information SI7). The LCA analysis also detected cicadas in the summer months, but that analysis assigned cicada sequences to Polyneurini, and the BLASTn re-analysis generally identified Graptopsaltria nigrofuscata. Thus, it appears that different species of cicada were detected using the two 16S markers. As in the 16S-epp analysis, the BLASTn re-analysis identified Largidae OTUs that had not been assigned to species in the LCA analysis as P. gutta. Within Lepidoptera, L. dispar was again identified in June, and no genus or species resolution was available for Erebidae (Supplementary Information SI7).
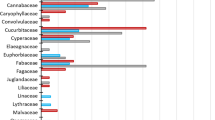
The NMDS plot of trnL rarefied relative read abundance data (sv = 0.16) indicated a seasonal difference in diet between spring and summer (Fig. 4A), which was supported by the results of the PerMANOVA (P < 0.01, F = 3.22, R2 = 0.24). The occurrence data also supported the presence of a seasonal difference in the NMDS plot (Fig. 4B; sv = 0.13) and PerMANOVA (P < 0.05, F = 1.64, R2 = 0.14). In spring, wood mouse diets were dominated by Fagaceae (Fig. 4C). Although the LCA analysis did not yield genus-level identification, nearly all observations with 100% identity in the DNA database belonged to Quercus, and thus we assumed all Fagaceae observations were of Quercus. Quercus variabilis dominates at the study site, with a minor proportion of Q. serrata (Nobukazu Nakagoshi, personal communication), and thus we assumed Q. variabilis to be the detected Fagaceae species. Further examination of the number of reads for this species showed that most of the reads for Fagaceae were derived from samples from early spring (93.6%; 76.2% in March and 17.4% in April; Supplementary Information SI8). In summer, mouse diets were dominated by unassigned Laurales, which we were unable to identify at a greater taxonomic resolution, and Euphorbiaceae (Malpighiales) (Fig. 4C). Given that the majority of hits in the DNA database for the latter belonged to Mallotus, we assumed that these reads belonged to Mallotus japonicus, as the genus is monotypic in Japan. Other OTUs detected at the genus level and frequently observed in summer included Lonicera (Caprifoliaceae), Rhododendron (Ericaceae), and Toxicodendron (Anacardiaceae) (Supplementary Information SI8). OTUs that were assigned to Ericaceae using LCA analysis were later confirmed as Rhododendron based on the BLASTn search re-analysis. Finally, Rutaceae OTUs were detected from three individuals in April, June, and August, albeit with a small number of reads (Supplementary Information SI8). Nearly all Rutaceae sequences detected with 100% identity in the DNA database belonged to a Citrus species.
Non-metric multidimensional scaling (NMDS) plot based on Bray–Curtis (A) and Jaccard (B) similarity indices for rarefied relative read data and occurrence data, respectively, obtained from trnL analysis. Open circles and open diamonds indicate samples obtained in spring and summer, respectively. (C) Read numbers assigned to each taxonomic category (orders and families) for trnL. Before plotting, the read numbers were rarefied and therefore normalized among samples to a pre-determined value for each marker as explained in the text. Reads associated with unassigned orders and Avena sp. were removed
Discussion
Dietary characteristics of A. speciosus on Innoshima Island and its ecological roles
The dietary components of A. speciosus, including both invertebrates and plants, shifted between spring (March–June) and summer (July–August) in our study area. Thus, the rainy season (Tsuyu), which runs from late June to late July, is likely a period of change in wood mouse diets. The rainy season is followed by a hot, humid summer in the study area, wherein various invertebrates become more active (e.g., cicadas; Hayashi and Saisho 2011). We found that wood mice consumed more Carabus beetles in July than any other surveyed month during the study (Fig. 5A), and they consumed large amounts of Platypleura (assumed to be P. kaempferi) and an unassigned cicada (assumed to be G. nigrofuscata) in July and August (Fig. 5F, H). Although only a minor diet component, wood mice consumed Vespula sp. in both July and August (Fig. 5B). Carabus has previously been reported as a diet item for A. speciosus in the Seto Inland Sea (Sato et al. 2019), but P. kaempferi, G. nigrofuscata, and Vespula sp. are novel dietary species. Given that A. speciosus is a terrestrial species, we suspect that larvae, exuviae, or dead individuals were the prey item, rather than live, mature adults of those flying species.
Monthly seasonal changes in dietary operational taxonomic units (OTUs). The vertical line indicates the number of rarefied reads per sample in each month. The most likely identity of each OTU (order, family, genus, or species) is shown above each panel along with the barcoding marker used: (A) Carabus – COI, (B) Vespula – COI, (C) Lymantria dispar – COI, (D) Sciaridae – COI, (E) Physopelta gutta – 16S-epp, (F) Platypleura – 16S-epp, (G) Largidae – 16S-kp, (H) Cicadidae – 16S-kp, (I) Quercus – trnL, (J) Rhododendron – trnL, (K) Laurales – trnL, and (L) Mallotus – trnL
For plants, broadly, consumption of a Fagaceae species (assumed to be Quercus variabilis) was high in early spring and then decreased toward the rainy season (Fig. 5I; Supplementary Information SI8). This observation suggests that wood mice may eat acorns preserved during the winter season. Although a direct observation of feeding on acorns is necessary to verify this hypothesis, the wood mouse may aid in the seed dispersal of Q. variabilis within its home range of ca. 500–2000 m2 each year (Oka 1992; Oishi et al. 2018). On the other hand, in summer, consumption of Rhododendron sp. increased in July, and an unassigned Laurales species was frequently detected in July and August (Fig. 5J, K). Mallotus sp. (assumed to be Mallotus japonicus) was more frequently consumed in August (Fig. 5L). In terms of the plant diet, the rainy season also appeared to be a turning point for changes in dietary habits.
Our findings suggest that A. speciosus may play an ecological role in pest control. We found that the wood mice ate Lymantria dispar, a cyclical-outbreak forest pest (Inoue et al. 2019), in June and July (Fig. 5C), suggesting that A. speciosus may suppress this forest pest by feeding on it, similar to the white-footed mouse Peromyscus leucopus in the USA (Elkinton et al. 1996). We also found that A. speciosus preyed on the gutta stink bug (P. gutta) and an unassigned Largidae species (assumed to be P. gutta) in March (Fig. 5E, G). Gutta stink bugs adversely affect citrus crops (Umeya and Okada 2003), and C. hassaku fruits are typically ripe in February and March on Innoshima Island. Predation by A. speciosus in March may therefore indicate a role of this mouse in reducing an orchard pest in the forest edge. The two cicada species we detected in the diet of A. speciosus, P. kaempferi and G. nigrofuscata, pierce fruit and consume tree sap at both the larval and adult stages (Umeya and Okada 2003). We also found that A. speciosus consumed Noctuoidea moth species in the families Erebidae, Lymantriidae, and Noctuidae, including many phytophagous moth species that could be harmful to orchard production (Umeya and Okada 2003), although we did not detect the notorious fruit-piercing moth species in Japan (Hattori 1969). In terms of the other moth species discussed, it is possible that Biston sp., which was abundant in A. speciosus feces according to the 16S-epp analysis, could have an adverse effect on citrus fruit production (Umeya and Okada 2003). Although the number of detected reads was low, we detected M. separata, which damages crops such as corn, and M. persicariae, which is a pest of soybean and Brassicaceae crops (Umeya and Okada 2003). We additionally detected a fly in the family Sciaridae in spring (Fig. 5D), which was assumed to be Bradysia sp. based on the BLASTn re-analysis. This genus is a pest of agricultural crops (Umeya and Okada 2003). We speculate that A. speciosus may perform an ecological role of suppressing the abundance of these pest species, similar to the common bent-wing bat (Miniopterus schreibersii) in the Mediterranean (Aizpurua et al. 2018). This potential role of A. speciosus in pest control should be tested in future studies that incorporate more intense sampling of wood mice.
The wood mice likely consumed some citrus fruit in April, June, and August, although at low abundance. Although this could be viewed negatively by orchard growers, we suggest that A. speciosus may play an even more important role in pest control as suggested above, highlighting the value of having forest ecosystems in proximity to agricultural operations.
Performance of three different barcoding markers for animal dietary profiling
In the present study, we performed multi-locus DNA metabarcoding analyses for animal diet profiling and recovered 230, 20, and 21 OTUs using the COI, 16S-epp, and 16S-kp barcoding markers, respectively. Our findings suggest that the COI primers developed by Zeale et al. (2011) were the most efficient markers for detecting invertebrate species represented in the diet of wood mice. The amplification of sequences from host wood mice (> 90% of the total reads) in the 16S-epp and 16S-kp analyses adversely affected the efficiency of these 16S markers. COI also outperformed the 16S genes in terms of the number of detected OTUs and taxonomic resolution in a study of bat diets (Tournayre et al. 2020), in which COI primers developed by Zeale et al. (2011) and 16S-epp primers developed by Epp et al. (2012) gave rise the highest and lowest numbers of invertebrate OTUs, respectively, among the 10 COI and two 16S barcoding markers. Nevertheless, even given this substantial difference in efficiency, we conclude that multi-locus DNA metabarcoding should be performed, because the three barcoding markers were complementary to one another in terms of detecting OTUs. In particular, Hemiptera was not detected using COI, but was found using 16S. Cicadidae (the two species of cicadas mentioned above) was characteristically found in the wood mouse diet in summer, whereas Largidae (stink bugs, a pest species) was observed in early spring. Therefore, the 16S markers proved to be useful for assessing seasonal dietary variation and the possible agricultural pest control ability of wood mice. Even the two 16S markers used in the present study varied in performance, as different cicada species were detected from different samples (Platypleura kaempferi from a sample in July with 16S-epp and Graptopsaltria nigrofuscata from samples in August with 16S-kp). We thus agree with the recommendation of previous studies to use multiple meta-barcoding markers (Alberdi et al. 2018; Browett et al. 2021), in contrast to Elbrecht et al. (2019). Because the detection of organisms is highly dependent on the sample contents (feces in this study), further case studies should be performed to determine which strategy with single or combined barcoding markers would be most effective for comprehensively assessing the diet of the wood mouse. Seasonal dietary analyses similar to those conducted in this study may be necessary because the efficiency of the primers used would change depending on the season and concomitant differences in dietary components.
Conclusions
Using the DNA metabarcoding approach, we demonstrated that a clear seasonal transition in diet occurred during the rainy season in the large Japanese wood mouse, A. speciosus. In addition, our results suggest the potential ecological role of this species in forest and agricultural pest management. Furthermore, we recommend multi-locus DNA metabarcoding analyses to achieve a comprehensive understanding of wood mouse diets. In future studies, we suggest the development of more efficient markers, the design of blocking primers targeting host DNA (especially for 16S markers), and the construction of a local DNA database (sequencing of the same markers used in this study for potential prey species collected in the field). These efforts would contribute to more precise clarification of the dietary components at the species and genus level and to improved understanding of the dynamics of diet shifts across seasons. In particular, with a local DNA database, one can potentially identify 23.8% of the reads for Noctuidae species and 77.9% of the reads for Lepidoptera that were not assigned to a lower taxonomic level and may include forest or agricultural pests.
Availability of data and material (data transparency)
We deposited genetic data obtained in this study to DDBJ (DNA Data Bank of Japan). The accession numbers for the sanger sequences are LC600288–LC600306. The accession number for the sequences produced by the next-generation sequencing is DRA011398. We also submit following supplementary data to support our findings.
References
Abe T, Oya T (1974) On the crop field murid rodents in Iwate. Bull Iwate Agric Exp Stan 18:23–29 ((in Japanese with English abstract))
Aizpurua O, Budinski I, Georgiakakis P, Gopalakrishnan S, Ibañez C, Mata V, Rebelo H, Russo D, Szodoray-Parádi F, Zhelyazkova V, Zrncic V, Gilbert MTP, Alberdi A (2018) Agriculture shapes the trophic niche of a bat preying on multiple pest arthropods across Europe: evidence from DNA metabarcoding. Mol Ecol 27:815–825
Alberdi A, Aizpurua O, Gilbert MTP, Bohmann K (2017) Scrutinizing key steps for reliable metabarcoding of environmental samples. Methods Ecol Evol 9:134–147
Alberdi A, Aizpurua O, Bohmann K, Gopalakrishnan S, Lynggaard C, Nielsen M, Gilbert MTP (2018) Promises and pitfalls of using high-throughput sequencing for diet analysis. Mol Ecol Resour 19:327–348
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Ando H, Mukai H, Komura T, Dewi T, Ando M, Isagi Y (2020) Methodological trends and perspectives of animal dietary studies by noninvasive fecal DNA metabarcoding. eDNA 2:391– 406.
Brockerhoff EG, Barbaro L, Castagneyrol B, Forrester DI, Gardiner B, González-Olabarria JR, Lyver PO’B, Meurisse N, Oxbrough A, Taki H, Thompson ID, van der Plas F, Jactel H, (2017) Forest biodiversity, ecosystem functioning and the provision of ecosystem services. Biodivers Conserv 26:3005–3035
Browett SS, Curran TG, O’Meara DB, Harrington AP, Sales NG, Antwis RE, O’Neill D, McDevitt AD (2021) Primer biases in the molecular assessment of diet in multiple insectivorous mammals. Mamm Biol 101:293–304
Corse E, Tougard C, Archambaud-Suard G, Agnèse J-F, Mandeng FDM, Bilong CFB, Duneau D, Zinger L, Chappaz R, Xu CCY, Meglécz E, Dubut V (2019) One-locus-several-primers: a strategy to improve the taxonomic and haplotypic coverage in diet metabarcoding studies. Ecol Evol 9:4603–4620
de Sousa LL, Silva SM, Xavier R (2019) DNA metabarcoding in diet studies: unveiling ecological aspects in aquatic and terrestrial ecosystems. eDNA 1:199–214.
Elbrecht V, Braukmann TWA, Ivanova NV, Prosser SWJ, Hajibabaei M, Wright M, Zakharov EV, Hebert PDN, Steinke D (2019) Validation of COI metabarcoding primers for terrestrial arthropods. PeerJ 7:e7745.
Elkinton J, Healy W, Buonaccorsi J, Boettner G, Hazzard A, Smith H (1996) Interactions among gypsy moths, white-footed mice, and acorns. Ecology 77:2332–2342
Epp LS, Boessenkool S, Bellemain EP, Haile J, Esposito A, Riaz T, Erséus C, Gusarov VI, Edwards ME, Johnsen A, Stenøien HK, Hassel K, Kauserud H, Yoccoz NG, Bråthen KA, Willerslev E, Taberlet P, Coissac E, Brochmann C (2012) New environmental metabarcodes for analyzing soil DNA: potential for studying past and present ecosystems. Mol Ecol 21:1821–1833
Hansen MC, Potapov PV, Moore R, Hancher M, Turubanova SA, Tyukavina A, Thau D, Stehman SV, Goetz SJ, Loveland TR, Kommareddy A, Egorov A, Chini L, Justice CO, Townshend JR (2013) High-resolution global maps of 21st-century forest cover change. Science 342:850–853
Hattori I (1969) Fruit-piercing moths in Japan. Japan Agricultural Research Quarterly 4:32–36
Hayashi M, Saisho Y (2011) The Cicadidae of Japan. Seibundo-Shinkosha (Tokyo, Japan) (In Japanese).
Hill SLL, Arnell A, Maney C, Butchart SHM, Hilton-Taylor C, Ciciarelli C, Davis C, Dinerstein E, Purvis A, Burgess ND (2019) Measuring forest biodiversity status and changes globally. Front for Glob Change 2:70
Hirota T, Hirohata T, Mashima H, Satoh T, Obara Y (2004) Population structure of the large Japanese field mouse, Apodemus speciosus (Rodentia: Muridae), in suburban landscape, based on mitochondrial D-Loop sequences. Mol Ecol 13:3275–3282
Inoue MN, Suzuki-Ohno Y, Haga Y, Aarai H, Sano T, Martemyanov VV, Kunimi Y (2019) Population dynamics and geographical distribution of the gypsy moth, Lymantria dispar, in Japan. Forest Ecol Manag 434:154–164
Kartzinel TR, Pringle RM (2015) Molecular detection of invertebrate prey in vertebrate diets: trophic ecology of Caribbean island lizards. Mol Ecol Resour 15:903–914
Mizushima S, Yamada E (1974) On the distribution and food habits of the murid rodents in agrosystems in Hokkaido. Jpn J Appl Entomol Zool 18:81–88 (in Japanese with English abstract)
Ohdachi SD, Ishibashi Y, Iwasa MA, Fukui D, Saitoh T (2015) The wild mammals of Japan, Second edition. Shoukadoh, Kyoto, Japan.
Oka T (1992) Home range and mating system of two sympatric field mouse species, Apodemus speciosus and Apodemus argenteus. Ecol Res 7:163–169
Oksanen J, Blanchet FG, Friendly M, Kindt R, Legendre P, McGlinn D, Minchin PR, O'Hara RB, Simpson GL, Solymos P, Stevens MHH, Szoecs E, Wagner H (2019) vegan: community ecology package. R package version 2.5–6. https://CRAN.R-project.org/package=vegan. Accessed 1 January 2021
Oishi K, Arakaki T, Nakamura M, Hata K, Sone K (2018) Spatial arrangement and size of home ranges of Apodemus speciosus inhabiting evergreen broad-leaved forest and adjacent cedar plantation, and migration between these stands. Mamm Sci (honyurui Kagaku) 58:23–31
R Core Team (2020) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. URL https://www.R-project.org/. Accessed 1 January 2021
Sato JJ, Kawakami T, Tasaka Y, Tamenishi M, Yamaguchi Y (2014) A few decades of habitat fragmentation has reduced population genetic diversity: a case study of landscape genetics of the large Japanese field mouse, Apodemus speciosus. Mamm Study 39:1–10
Sato JJ (2016) A review on the process of mammalian faunal assembly in Japan—insight from the molecular phylogenetics. In: Motokawa M and Kajihara H (eds) Species Diversity of Animals in Japan. Springer Japan, Tokyo, pp 49–116.
Sato JJ, Tasaka Y, Tasaka R, Gunji K, Yamamoto Y, Takada Y, Uematsu Y, Sakai E, Tateishi T, Yamaguchi Y (2017) Effects of isolation by continental islands in the Seto Inland Sea, Japan, on genetic diversity of the large Japanese field mouse, Apodemus speciosus (Rodentia: Muridae), inferred from the mitochondrial Dloop region. Zool Sci 34:112–121
Sato JJ, Shimada T, Kyogoku D, Komura T, Uemura S, Saitoh T, Isagi Y (2018) Dietary niche partitioning between sympatric wood mouse species (Muridae: Apodemus) revealed by DNA meta-barcoding analysis. J Mammal 99:952–964
Sato JJ, Kyogoku D, Komura T, Maeda K, Inamori C, Yamaguchi Y, Isagi Y (2019) Potentials and pitfalls of the DNA metabarcoding analyses for the dietary study of the large Japanese wood mouse Apodemus speciosus on Seto Inland Sea islands. Mamm Study 44:221–231
Sato JJ, Aiba H, Ohtake K, Minato S (2020) Evolutionary and anthropogenic factors affecting the mitochondrial D-loop genetic diversity of Apodemus and Myodes rodents on the northern slope of Mt. Fuji Mamm Stud 45:315–325
Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E (2007) Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res 35:e14.
Tachibana K, Ishii N, Kato S (1988) Food habits of small mammals during a sawfly (Pristifora erichsoni Hartig) outbreak. J Jap Forestry Soc 70:525–528
Tanabe AS, Toju H (2013) Two new computational methods for universal DNA barcoding: a benchmark using barcode sequences of bacteria, archaea, animals, fungi, and land plant. PLoS ONE 8: e76910.
Tatsukawa K, Murakami O (1976) On the utilization of the Japanese wood mouse Apodemus speciosus (Mammalia: Muridae). Physiol Ecol Jpn 17:133–144 (in Japanese with English Abstract)
Tournayre O, Leuchtmann M, Filippi-Codaccioni O, Trillat M, Piry S, Pontier D, Charbonnel N, Galan M (2020) In silico and empirical evaluation of twelve metabarcoding primer sets for insectivorous diet analyses. Ecol Evol 10:6310–6332
Umeya K, Okada T (2003) Agricultural insect pest in Japan. Zenkoku Nouson Kyouiku Kyoukai, Japan (in Japanese).
Vamos E, Elbrecht V, Leese F (2017) Short COI markers for freshwater macroinvertebrate metabarcoding. Metabarcoding Metagenom 1:e14625.
Zahiri R, Kitching IJ, Lafontaine JD, Mutanen M, Kaila L, Holloway JD, Wahlberg N (2010) A new molecular phylogeny offers hope for a stable family level classification of the Noctuoidea (Lepidoptera). Zool Scr 40:158–173
Zeale MR, Butlin RK, Barker GL, Lees DC, Jones G (2011) Taxon-specific PCR for DNA barcoding arthropod prey in bat faeces. Mol Ecol 11:236–244
Acknowledgements
We thank Nobukazu Nakagoshi for identification of the Fagaceae plant species from the acorns that we collected in the survey location. We are also grateful to Tomoyasu Shirako for his help in bioinformatics analyses of the trnL sequence and Kouki Yasuda for his help in making the map figure by QGIS.
Funding
This work was supported by the research fund from the Private University Research Branding Project (Ministry of Education, Culture, Sports, Science and Technology, Japan) and the Green Science Research Center in Fukuyama University (granted to Jun J. Sato).
Author information
Authors and Affiliations
Contributions
JJS planned and financially supported the research project. JJS, YO, and NN were involved in the sampling of Apodemus speciosus from the Innoshima Island in the Seto Inland Sea, Japan. YO, NN, and KM undertook the laboratory work. JJS conducted the analyses. JJS drafted the manuscript. All authors reviewed the manuscript and joined the discussion.
Corresponding author
Ethics declarations
Ethics approval (include appropriate approvals or waivers)
We obtained permission from Hiroshima Prefecture to collect the wood mice samples in the Innoshima island (no approval code from the prefecture) and conducted protocols approved by the Animal Care and Use Committee of Fukuyama University (approval code: H30-Animal-8). We followed the guidelines of the Procedure of Obtaining Mammal Specimens established by the Mammal Society of Japan (https://www.mammalogy.jp/en/guideline.pdf; accessed 1 June 2021).
Consent to participate (include appropriate statements)
All authors contributed to the study as described in the author’s contribution above, read the manuscript, and discussed the contents.
Consent for publication (include appropriate statements)
All authors agreed with the submission and publication of this manuscript in Mammal Research.
Competing interests (include appropriate disclosures).
The authors declare no competing interests.
Additional information
Communicated by: Cino Pertoldi
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13364_2021_601_MOESM2_ESM.pptx
Supplementary file2 Supplementary Information SI2.— Rarefaction curves obtained for each molecular marker. (PPTX 415 KB)
13364_2021_601_MOESM3_ESM.pptx
Supplementary file3 Supplementary Information SI3.— Rarefied read numbers assigned to each taxonomic category (orders and families) for COI. (PPTX 72 KB)
13364_2021_601_MOESM4_ESM.xlsx
Supplementary file4 Supplementary Information SI4.— Rarefied reads for the detected OTUs in the COI analysis (XLSX 40 KB)
13364_2021_601_MOESM5_ESM.xlsx
Supplementary file5 Supplementary Information SI5.— Rarefied reads for the detected OTUs in the 16S-epp analysis(XLSX 20 KB)
13364_2021_601_MOESM6_ESM.pptx
Supplementary file6 Supplementary Information SI6.—NMDS plot of 16S-kp including outlier samples (#13 and #14) (PPTX 120 KB)
13364_2021_601_MOESM7_ESM.xlsx
Supplementary file7 Supplementary Information SI7.— Rarefied reads for the detected OTUs in the 16S-kp analysis (XLSX 19 KB)
13364_2021_601_MOESM8_ESM.xlsx
Supplementary file8 Supplementary Information SI8.— Rarefied reads for the detected OTUs in the trnL analysis(XLSX 22 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Sato, J.J., Ohtsuki, Y., Nishiura, N. et al. DNA metabarcoding dietary analyses of the wood mouse Apodemus speciosus on Innoshima Island, Japan, and implications for primer choice. Mamm Res 67, 109–122 (2022). https://doi.org/10.1007/s13364-021-00601-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13364-021-00601-7