Abstract
A comparative study on the proton-bound complexes of 1-methylcytosine (1-mC) with 1-methylguanine (1-mG) and 9-methylguanine (9-mG), [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+, respectively, was carried out using energy-resolved collision-induced dissociation (ER-CID) experiments in combination with quantum chemical calculations. In ER-CID experiments, the measured survival yields indicated an essentially identical stability for the two proton-bound complexes. In comparison with the lowest-energy structures and base-pairing energetics predicted at the B3LYP/6-311+G(2d,2p) theory level, both complexes produced in this study were suggested to be proton-bound Hoogsteen base pairs. Curiously, despite the similarity in structures, binding energetics, and potential energy surfaces predicted by the B3LYP theory, the fragment branching ratios exhibited an intriguing alternation between the two proton-bound Hoogsteen base pairs. The CID of [1-mC:1-mG:H]+ produced protonated cytosines, [1-mC:H]+, more abundantly than [1-mG:H]+, whereas that of [1-mC:9-mG:H]+ gave rise to a more pronounced production of protonated guanines, [9-mG:H]+. However, using the proton affinities of moieties predicted by the high-accuracy methods, including CBS-QB3 and the Guassian-4 theory, the anomaly known for [Cytosine:Guanine:H]+ (J. Am. Soc. Mass Spectrom. 29, 2368–2379 (2018)) successfully accounted for the alternated branching ratios. Thereby, the anomaly, more specifically, the production of proton-transferred fragments of O-protonated cytosines in the CID of proton-bound Hoogsteen base pairs, is indeed real, which is disclosed as the alternated branching ratios in the CID spectra of [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+ in this study.

.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mass spectrometry has provided a powerful means to elucidate structures and base-pairing energy of non-covalent complexes of proton-bound nucleic acid bases, which provide structural motifs to form altered DNA structures upon protonation. For example, infrared multiple photon dissociation (IRMPD) spectroscopy [1,2,3,4] successfully probed the triply bound nature of proton-bound base pairs of cytosines [5,6,7]. This has deepened our understanding of base-pairing interactions involved in tetrameric DNA structures arising from cytosine-rich oligomers in the acidic conditions [8,9,10,11], which are known as DNA i-motifs [12]. In addition, energy-resolved collision-induced dissociation (ER-CID) experiments in single or multiple collision conditions have offered an accurate way to determine binding energetics for proton-bound base pairs of cytosines and deoxycytidines along with thermochemistry such as the proton affinities (PA) of their moieties [13,14,15,16,17,18]. IRMPD spectroscopy was also employed to examine various proton-bound complexes of nucleic acid bases including adenine (A) dimers, [A:A:H]+ [19], heterodimers of cytosine (C) and guanine (G), [C:G:H]+ [20], and for metal cation-bound complexes such as G-tetrads [21].
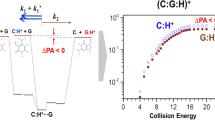
In particular, a recent IRMPD spectroscopy elucidated the structures of proton-bound heterodimers of [C:G:H]+ produced by electrospray ionization (ESI) for the first time in the gas phase [20]. The spectroscopic investigation along with differential ion mobility spectrometry (DIMS) revealed an interesting change in conformational preference, which depended on the pH of sample solutions (methanol:water = 1:1). In the study, ESI preferentially produced proton-bound Hoogsteen base pairs in acidic conditions (pH = 3.2; 66% Hoogsteen), whereas protonated Watson–Crick (WC) base pairs were found to be dominant in non-acidified conditions (pH = 5.8; 91% WC) (Figure 1).
In fact, proton-bound Hoogsteen base-pairing is another important cause of altered DNA structures by protonation [22]. Although WC base-pairing is stable at physiological conditions, proton-bound Hoogsteen base-pairing is known to occur transiently under certain circumstances [23,24,25,26,27,28,29], wherein N3-protonated C binds to the Hoogsteen edge of G in WC base pairs. This leads to the formation of triplet, [C:H+•••G:C], and triplexes of nucleotide oligomers [28, 29]. Such formation of triple stranded DNA structures via a Hoogsteen interaction may be related to human diseases such as Friedreich’s ataxia [30, 31]. In addition, the transient formation of the Hoogsteen strand in DNA double helixes may have a role in expanding the genetic code beyond the capacity served only by WC base-pairing [25,26,27].
One theoretical study predicted that the proton-bound Hoogsteen base pair is the most stable conformation for isolated [C:G:H]+ and thus would be dominant in the gas phase [32]. The base-pairing in proton-bound Hoogsteen base pairs utilizes two hydrogen bonds, of which one is an ionic hydrogen bond offered by an extra proton attached to the N3-position of the C moiety [33]. The strength of interaction involved in the Hoogsteen base-pairing in [C:G:H]+ was predicted to be rather strong at 159.87 kJ/mol [34], which is stronger than the theoretical binding energy of neutral G:C WC base pairs at 99.6 kJ/mol [35]. It is still weaker than that of triply hydrogen-bonded C dimers, [C:C:H]+, at 171.7 and 169.9 kJ/mol, which are theoretical and experimental values, respectively [14]. In the Hoogsteen base pair, the proton transfer (PT) in the ionic hydrogen bond between the two moieties may be facile due to the low predicted energy barrier for the PT of 4.90 kJ/mol (ΔE0PT) [33].
In the CID experiments, the proton-bound Hoogsteen base pair of [C:G:H]+ exhibited an intriguing behavior in the fragment abundance ratio [36]. In view of the kinetic method [37,38,39], it is generally expected that the protonated moiety of a larger PA would be produced more abundantly in the CID of a proton-bound complex of [A•••H+•••B] under multiple collision conditions. However, it was found that the CID of [C:G:H]+ had a more pronounced formation of protonated C, [C:H]+, than [G:H]+ despite the smaller experimental PA of C (949.8 kJ/mol) than that of G (959.4 kJ/mol) [40, 41]. To understand the anomalous CID behaviors of [C:G:H]+, ER-CID experiments in combination with a theoretical study on dissociation pathways for a homolog series of proton-bound Hoogsteen base pairs, [C:G:H]+, [1-methylcytosine (1-mC):G:H]+, and [5-methylcytosine:G:H]+, were carried out [34]. In the study, an additional fragmentation pathway that involves proton transfer during dissociation was suggested to be available, which leads to the generation of proton-transferred products, O2-protonated cytosines, in addition to N3-protonated cytosine fragments from direct dissociation. The presence of the additional fragmentation channel for [C:H]+ fragments was considered as the cause of the anomaly, which was the observation of more abundant [C:H]+ fragments than [G:H]+ in the CID of proton-bound Hoogsteen base pair of [C:G:H]+.
In a continuing effort to understand the anomaly, we further investigated the ER-CID behaviors of proton-bound Hoogsteen base pairs of [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+ in a comparative study. As a result, this study demonstrates that the anomaly, i.e., the proton transfer occurring in the course of the CID of proton-bound Hoogsteen base pairs, is indeed present and determines the apparent CID behaviors.
Experimental and Theoretical Methods
Materials
Chemicals, including cytosine (C), 1-methylguanine (1-mG), and 9-methylguanine (9-mG), were commercially obtained from Sigma–Aldrich Korea (Suwon, Korea). 1-methylcytosine (1-mC) was purchased from Synchem UG & Co. KG (Felsberg, Germany). Other chemicals including acetonitrile and acetic acid were also obtained from Sigma–Aldrich Korea. The chemicals were used without further purification.
ER-CID Experiments
ER-CID experiments were performed on an ion trap mass spectrometer (LTQ XL, Thermo Scientific, MA, USA). Proton-bound base pairs were produced by using the ESI of binary sample solutions (200 μM each) prepared in a 1:1 solution of water and acetonitrile with 1% acetic acid (pH = 3.3). The sample solutions were electrosprayed by direct infusion at a flow rate of 10 μl/min. The temperature of inlet capillary was generally set to 150 °C. Precursor ions, which were proton-bound base pairs, stored in the linear ion trap were excited for 30 msec, during which the ions underwent CID by multiple collisions with the collision gas, He, at about 1 mTorr. For the ER-CID experiments, the excitation energy was set in terms of the normalized collision energy (NCE), the instrument’s parameter in the arbitrary unit (a.u.), of which the scan step was chosen to be 0.5 or 1. The survival yield (S.Y.) for the proton-bound dimers was monitored in terms of the ratio of the abundance of surviving precursor ions, I(p), to the sum of abundances of all ions, Itotal, which is the sum for all fragments (f), Ii(f), and surviving precursor ions (p), I(p), measured in the MS/MS spectra; S.Y. = I(p)/Itotal = I(p)/(ΣIi(f) + I(p)). Similarly, the formation yield (F.Y.) for a fragment was defined as F.Y. (f) = I(f)/Itotal. The yields were obtained as a function of NCE. Using the least square method, the S.Y. curves were fitted to a relation, S.Y. = [1+ (NCE/CID50)n]−1. The fitted value of CID50 corresponds to the collision energy at which 50% depletion of precursor ions occurs, which represents the relative stability of precursor ions. The value n represents the steepness of the declining region of the fitted curve [42]. The ER-CID data were obtained and averaged for three independent measurements, of which standard deviations were measured to be very small and comparable to the sizes of the symbols depicted in the graphs. For CID behaviors, the influences of different solvent composition (methanol:water = 1:1), pH (pH = 7.3 – 7.6) and temperature of inlet capillary (120–400 °C) were also examined.
Quantum Chemical Calculations
A computational study was performed using the Gaussian 09 program suite [43]. Density functional theory using the B3LYP hybrid functional has been widely utilized in many theoretical investigations of nucleic acid bases and their base pairs. In particular, the theory level of B3LYP/6-311+G(2d,2p) was found to be very reliable in predicting the structures and base-pairing interactions of proton-bound nucleic acid base pairs in comparison with data obtained by threshold CID (TCID) experiments [14, 15]. Thus, this computational study employed the B3LYP/6-311+G(2d,2p) theory in exploring the potential energy surfaces for the proton-bound base pairs of [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+, which includes the localization of minimum energy structures, transition states (TS), and intermediates (INT), as well as the calculation of related energetics. For vibration frequency calculations, a scale factor of 0.9804 was used. The calculations further predicted thermochemical quantities such as enthalpy (H) and Gibbs energy (G) at 298.15 K. As for the interaction energy between the two moieties in a base pair, [A:H+•••B], the dissociation energy with zero-point energy correction defined as D0 = [E0(A:H+) + E0(B)]−E0([A:H+•••B]) was evaluated. The dissociation energy was further corrected for basis set superposition errors (D0,BSSE) using the relaxed counterpoise method [44]. Polarizable continuum model (PCM) calculations were carried out when the energetics in the aqueous environment was examined [45]. In searching for TS, a potential energy scan was performed for the areas of interest on potential energy surfaces. TS was then localized using QST3 calculations [46], which were further examined by vibration frequency calculations.
To predict the energetics, including the PAs of monomers, more reliably, high-accuracy methods, including the Gaussian-4 theory (G4) and its variations (G4MP2) [47, 48], as well as the complete basis set method (CBS-QB3), were utilized [49, 50]. The high-accuracy methods are known for providing high accuracy in predicting the thermochemistry of molecules from benchmarking studies [47,48,49,50,51,52,53], which is comparable to the chemical accuracy (1 kcal/mol). The high-accuracy methods are computationally expensive, in particular, G4, for such large systems as the proton-bound base pairs of the present study. Thus, potential energy surfaces were explored at the theory level of B3LYP/6-311+G(2d,2p) [14, 15], which is ss computationally demanding than the high-accuracy methods.
Data and Results
Alternation in Branching Ratios in ER-CID of [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+
ER-CID experiments were carried out for the proton-bound heterodimers of 1-mC with 1-mG and 9-mG, which were produced by the ESI of the acidic solutions in this study (pH = 3.3). Figure 2 displays the S.Y. curves for the proton-bound precursor ions of [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+, which also display that for [1-mC:C:H]+ measured under the same condition for comparison. In general, the S.Y. curves revealed relative stabilities for the precursor ions. The observed order in energy (NCE) was [1-mC:C:H]+ (CID50 = 10.40) > [1-mC:9-mG:H]+ (9.62) ≈ [1-mC:1-mG:H]+ (9.57). In the S.Y. curves, it is evident that the two base pairs, [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+, possess an essentially identical stability with respect to CID by multiple collisions. It is also shown that their stabilities are still weaker than the triply bound complex of [1-mC:C:H]+ involving triple hydrogen bonds, of which the interaction energy was measured to be 166.6 kJ/mol by the TCID experiments [16]. The comparable stability suggests that both proton-bound base pairs, which differ only in the methylation positions of the G moieties, may be formed by the same type of base-paring interaction, presumably.
The F.Y. curves obtained for [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+ by the ER-CID experiments are presented in Figure 3. Interestingly, the F.Y. curves display distinctly different CID behaviors for the two proton-bound base pairs. It is noticeable that the production of the protonated C moiety, [1-mC:H]+, was pronounced in the CID of [1-mC:1-mG:H]+, whereas the protonated G moiety, [9-mG:H]+, was more abundant than [1-mC:H]+ in the CID spectra of [1-mC:9-mG:H]+. The branching ratios were measured to be 0.8:0.2 for [1-mC:H]+:[1-mG:H]+ in the CID of [1-mC:1-mG:H]+ and 0.4:0.6 for [1-mC:H]+:[9-mG:H]+ in that of [1-mC:9-mG:H]+, approximately. Since it appears that the only difference between the two base pairs is the methylation sites in 1-mG and 9-mG, the observed alternation in the branching ratios is curious.
In the previous IRMPD study, the conformational preference of [C:G:H]+ between proton-bound Hoogsteen and WC isomers was found to vary depending on the pH of ESI sample solutions [20]. In this regard, the effects of different solvent composition, pH of sample solutions, and temperature of inlet capillary of the mass spectrometer on the CID behaviors of both complexes were examined. However, there were no noticeable influences on the CID curves, particularly on the branching ratios, for the proton-bound base pairs (Figures 1S and 2S in the supporting information). It suggests that the proton-bound complexes produced in this study can be represented by a predominant conformation, which is mostly likely the most thermodynamically stable isomers of the proton-bound complexes.
Lowest-Energy Structures of [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+: Proton-Bound Hoogsteen Base Pairs
To understand the alternation observed in the branching ratios of fragments produced by the CID of [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+, we performed a theoretical study for the proton-bound base pairs at the theory level of B3LYP/6-311+G(2d,2p) [14, 15]. In search for plausible structures that might represent the conformations of precursor ions in the gas phase, extensive exploration of dimer structures was carried out in the same way as utilized in the previous studies [32, 34]. In this study, for protonated moieties, the protonation of the N3- and (cis-) O2-positions of 1-mC and the N7-position of 1-mG and 9-mG moieties was considered (Figure 3S). The lowest-energy structures found for [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+, five conformers for [1-mC:1-mG:H]+, and four for [1-mC:9-mG:H]+ within ΔG < 25 kJ/mol are given in Fig. 4S and 5S with their predicted energies in Tables 1S and 2S.
In the results, it is to be noted that the lowest-energy structures for both proton-bound dimers are proton-bound Hoogsteen base pairs, in which two hydrogen bonds, including an ionic hydrogen bond, stabilize the proton-bound dimers (Figure 4). For [1-mC:1-mG:H]+, the proton-bound Hoogsteen base pair (I) was predicted to be more stable than the second stable structure (II) by 8.37 kJ/mol in the gas phase (ΔG) and 5.40 kJ/mol in the aqueous environment (ΔGPCM). As for [1-mC:9-mG:H]+, the Hoogsteen base pair (I) was more stable than the next stable one (II) by 10.99 and 10.29 kJ/mol in ΔG and ΔGPCM, respectively. In view of the Boltzmann population, the ΔG value of 8 kJ/mol accounts for more than 95% of the population in the gas phase in thermal equilibrium. As the proton-bound Hoogsteen base pairs may also be more stable in the sample solution (ΔGPCM), the theoretical results suggest that the proton-bound Hoogsteen base pairs are most likely the gas-phase conformations for [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+ produced by ESI in this study.
In addition, the dissociation energies (D0,BSSE) for proton-bound Hoogsteen base pairs, [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+, to their respective lowest-energy product states, [1-mG:H]+ + 1-mC and [9-mG:H]+ + 1-mC, were calculated to be 149.58 and 153.94 kJ/mol, respectively (Table 1). The difference in D0,BSSE is 4.36 kJ/mol, which is a small difference that agrees well with the comparable stability of [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+ observed in the S.Y. curves. These suggest that proton-bound Hoogsteen base pairs represent both the proton-bound complexes of [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+ produced in the gas phase under our experimental conditions.
As for the possible presence of protonated WC base pairs [20], the WC conformation for [1-mC:9-mG:H]+ was located high in energy (ΔE0 = 16.49, ΔG = 18.37 kJ/mol; IV in Figure 5S) in this theoretical study, of which the lowest dissociation energy was calculated to be 137.23 kJ/mol (D0,BSSE; Table 2S). As for the [1-mC:1-mG:H]+ base pairs, due to the methylation at the N1-position of guanine, WC base-pairing is not available for this complex. The dissociation energy for the protonated WC base pair for [1-mC:9-mG:H]+ is significantly different than that of the Hoogsteen base pair of [1-mC:1-mG:H]+ (149.58 kJ/mol), which does not account for the comparable stability for the two base pairs evident in the S.Y. curves. In addition, as previously found in the IRMPD spectroscopy, the acidic sample conditions for ESI under our experimental conditions may prefer to produce proton-bound Hoogsteen base pairs [20, 34]. As a result, proton-bound WC base pairs were not considered in this study. Considering the absence of noticeable influences of solvent, pH, and temperature on the CID behaviors (Figures 1S and 2S), the proton-bound complexes produced in our conditions are mostly likely the most stable and thus predominant conformers of the complexes, for which this theoretical study predicted proton-bound Hoogsteen base pairs. In other words, the effects of other isomers may not be significant for the experimental results in this study.
Three Fragmentation Pathways for Proton-Bound Hoogsteen Base Pairs
We further explored fragmentation pathways for the proton-bound Hoogsteen base pairs of [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+ at the B3LYP/6–311+G(2d,2p) theory level. It is to be noted that the resulting potential energy diagrams for the two proton-bound base pairs are essentially isomorphic to each other and are similar to the previous one for [C:G:H]+ [34] (Figure 5). However, this theory level was not successful in locating the transition state, TSPT(O2), for [1-mC:1-mG:H]+. Instead, TSPT(O2)† was located using a slightly different basis set (†) of 6–311+G(2d,p).
Potential energy diagrams for (a) [1-mC:1-mG:H]+ and (b) [1-mC:9-mG:H]+ with predicted energy differences from the lowest-energy structures in parenthesis (ΔEe/ ΔE0/ ΔH/ ΔG at 298.15 K; kJ/mol) at the B3LYP/6-311+G(2d,2p) theory level. (†) denotes the energy obtained by B3LYP/6-311+G(2d,p). The structures of INT and TS are given in Figure 4S. (ΔPA = PA(1-mC)–PA(x-mG))
The potential energy diagrams indicate that three pathways are energetically accessible for the dissociation of proton-bound Hoogsteen base pairs. Fragmentations leading to [1-mC(N3):H]+ and [x-mG:H]+ are the channels that are typically considered as the two pathways competing for the proton of the ionic hydrogen bond in a proton-bound dimer in the kinetic method. However, due to the low energy barriers for TSPT(O2), dissociation into proton-transferred products of [1-mC(O2):H]+ may also be facile in the CID of proton-bound Hoogsteen base pairs. The presence of an additional pathway giving rise to the proton-transferred fragments of [1-mC(O2):H]+ would increase the apparent abundance of [1-mC:H]+ in m/z, which was suggested as the cause for the anomalous CID behaviors of [C:G:H]+ [34].
In fact, it is also to be noted that the dissociation of proton-bound, triply bound dimers of [1-mC:1-mC:H]+ by IRMPD induced by the irradiation of a CO2 laser led to the production of predominant [1-mC(O2):H]+ fragments [1]. This indicates that proton transfer is facile, even in the dissociation of triply hydrogen-bonded base pairs by the slow heating mechanism. Thus, the competition among three fragmentation pathways leading to the production of [1-mC(N3):H]+, [1-mC(O2):H]+, and [x-mG:H]+ fragments may play a key role in determining apparent CID behaviors.
In the theoretical study at the B3LYP/6-311+G(2d,2p) theory level, the H of protonation (ΔHp; 298.15 K), which corresponds to PA, for 1-mC were predicted to be 965.76 and 964.79 kJ/mol when the N3- and O2-positions of 1-mC are protonated, respectively (Table 2). This agrees well with the experimental reports of 971 kJ/mol obtained by the kinetic method [54] and 964.7 kJ/mol obtained by TCID experiments [16]. The ΔHp for 1-mG and 9-mG were calculated to be 968.02 and 973.46 kJ/mol in this study, respectively. Thus, this B3LYP study predicted larger PAs for methylated guanine moieties than those of 1-mC; 9-mG > 1-mG > 1-mC(N3) > 1-mC(O2), wherein the protonation site of 1-mC is indicated in parenthesis. In fact, the PAs of moieties are the energy values that determine the relative energetics of the product states. For both base pairs, the B3LYP theory predicted ΔPA < 0 (= PA(1-mC)–PA(x-mG)), which makes the potential energy diagrams essentially isomorphic (Figure 5).
Alternated Branching Ratios by Anomaly
However, despite the resemblance in potential energy surfaces explored by the B3LYP theory, the CID of the proton-bound Hoogsteen base pairs of this study, [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+, exhibited an intriguing alternation in branching ratios. In fact, it is generally expected from previous benchmarking tests that the accuracy of B3LYP theory without dispersion corrections for non-covalent interactions may not be better than 3 kcal/mol [55, 56].
To address this issue more carefully, we examined the PAs of monomers by using high-accuracy methods, including CBS-QB3, G4MP2, and G4. Those are the compound methods developed to provide a high accuracy in predicting the thermochemical properties of molecules, which approaches the chemical accuracy (1 kcal/mol) [47,48,49,50,51,52,53]. The predicted thermochemistry for protonation, ΔHp and ΔGp, along with their averages are given in Table 2.
The predicted values by the high-accuracy methods were found to be very close. The standard deviations for the values obtained by the three different methods ranged only between 1.2–1.6 kJ/mol, whereas the values predicted by the B3LYP theory were quite deviated from the high-accuracy values. For example, the B3LYP theory predicted larger ΔHp values by 1.74–9.01 kJ/mol than the average values of three high-accuracy methods, which far outlie the standard deviations for the three methods. More importantly, the three high-accuracy methods equivocally predicted a different order in energy for protonation from that of the B3LYP theory of 9-mG > 1-mG > 1-mC(N3) > 1-mC(O2). The order in ΔHp predicted by the high-accuracy methods was 9-mG > 1-mC(O2) > 1-mG > 1-mC(N3), which as a consequence altered the energetics of the product states significantly (Figure 6).
Potential energy diagrams for (a) [1-mC:1-mG:H]+ and (b) [1-mC:9-mG:H]+ with predicted energy differences using the average values obtained by the high-accuracy methods in parenthesis (ΔEe/ΔE0/ΔH/ΔG at 298.15 K; kJ/mol). Dissociation energies (D0, ΔH, ΔG; kJ/mol) are values obtained by using the CBS-QB3 theory. ΔHp and ΔGp (298.15 K) predicted at other theory levels are given in Table 2. (ΔPA = PA(1-mC)–PA(x-mG))
The improved product states in energetics for the two proton-bound base pairs are not isomorphic any more. It is evident in the potential energy diagrams that the formation of the proton-transferred products of [1-mC(O2):H]+ provides a more energetically favorable pathway for the dissociation of [1-mC:1-mG:H]+, whereas the pathway leading to the production of [9-mG:H]+ is more favorable in the fragmentation of [1-mC:9-mG:H]+. In addition, the CBS-QB3 theory predicted the dissociation energies (D0) into the lowest-energy product state to be 178.96 and 176.72 kJ/mol for [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+, respectively. The difference in D0 is only 2.24 kJ/mol, which is even smaller than the predicted difference by the B3LYP theory of 4.36 kJ/mol. It thus more clearly accounts for the comparable stability observed for the proton-bound precursor complexes in the ER-CID experiments. It is also to be noted that the predicted ΔHps for C and G also altered the product states of the previous [C:G:H]+ [34], which now resembles that of [1-mC:1-mG:H]+. In both cases of [C:G:H]+ and [1-mC:1-mG:H]+, the anomaly, that is production of more abundant protonated cytosine fragments, [C:H]+ and [1-mC:H]+, respectively, was evident.
If the three fragmentation pathways are assumed to be independent, one can apply the simple relation of the kinetic method [37, 38], ln(kA/kB) = ln([A:H+]/[B:H+]) ≈ ΔΔHp/RTeff, to estimate the branching ratios in the CID of the proton-bound dimers of [A•••H+•••B]. Using the average ΔHps for moieties obtained by the high-accuracy methods (Table 2) and Teff = 300 K, arbitrarily, the apparent branching ratios were estimated to be [1-mC:H]+:[1-mG:H]+ = 0.85:0.15 for [1-mC:1-mG:H]+ ([1-mC(N3):H]+:[1-mC(O2):H]+:[1-mG:H]+ = 0.17:1:0.20) and [1-mC:H]+:[9-mG:H]+ = 0.29:0.71 for [1-mC:9-mG:H]+ ([1-mC(N3):H]+:[1-mC(O2):H]+:[9-mG:H]+ = 0.06:0.35:1). This accounts for the observed alternation in the branching ratios of 0.8:0.2 and 0.4:0.6 for [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+, respectively. However, we cannot completely exclude the effects that may be caused by the presence of other isomers and the entropy differences between fragmentation pathways, although the results of this study show that those effects may not be significantly involved in these cases of CID of [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+.
Accordingly, the anomaly, i.e., the production of proton-transferred fragments, [1-mC(O2):H]+, is disclosed by the observed alternation in the branching ratios for the CID of [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+.
Conclusions
This study demonstrates that understanding the dissociation pathways is crucial in determining accurate thermochemical properties of moieties in proton-bound complexes based on the kinetic method, particularly when multiple hydrogen bonding is involved. In addition, although the B3LYP theory is still reliable in predicting the structures and base-pairing interactions of proton-bound base pairs, theories that have a higher accuracy need to be considered when dealing with a small and critical energy difference such as ΔΔHp between the cytosine and guanine moieties. This, in fact, determined the dissociation behaviors of proton-bound Hoogsteen base pairs in the present study.
In conclusion, the alternated branching ratios in CID of [1-mC:1-mG:H]+ and [1-mC:9-mG:H]+ indicate the anomaly, which indeed arises from the presence of proton transfer reactions occurring in the course of the dissociation of proton-bound Hoogsteen base pairs.
References
Oomens, J., Moehlig, A.R., Morton, T.H.: Infrared multiple photon dissociation (IRMPD) spectroscopy of the proton-bound dimer of 1-methylcytosine in the gas phase. J. Phys. Chem. Lett. 1, 2891–2897 (2010)
Ung, H.U., Moehlig, A.R., Kudla, R.A., Mueller, L.J., Oomens, J., Berden, G., Morton, T.H.: Proton-bound dimers of 1-methylcytosine and its derivatives: vibrational and NMR spectroscopy. Phys. Chem. Chem. Phys. 15, 19001–19012 (2013)
Yang, B., Wu, R.R., Berden, G., Oomens, J., Rodgers, M.T.: Infrared multiple photon dissociation action spectroscopy of proton-bound dimers of cytosine and modified cytosines: effects of modifications on gas-phase conformations. J. Phys. Chem. B. 117, 14191–14201 (2013)
Kwon, S., Oh, H.B., Han, S.Y.: Infrared multiple photon depletion of the gas-phase proton-bound cytosine dimer. Chem. Lett. 44, 1756–1758 (2015)
Šponer, J., Leszczynski, J., Vetterl, V., Hobza, P.: Base stacking and hydrogen bonding in protonated cytosine dimer: the role of molecular ion-dipole and induction interactions. J. Biomol. Struct. Dyn. 13, 695–706 (1996)
Han, S.Y., Oh, H.B.: Theoretical study of the ionic hydrogen bond in the isolated proton-bound dimer of cytosine. Chem. Phys. Lett. 432, 269–274 (2006)
Lieblein, A.L., Krämer, M., Dreuw, A., Fürtig, B., Schwalbe, H.: The nature of hydrogen bonds in cytidine H + cytidine DNA base pairs. Angew. Chem. Int. Ed. 51, 4067–4070 (2012)
Gehring, K., Leroy, J.-L., Guéron, M.: A tetrameric DNA structure with protonated cytosine–cytosine base pairs. Nature. 363, 561–565 (1993)
Darlow, J.M., Leach, D.R.F.: Secondary structure in d(CGG) and d(CCG) repeat tracts. J. Mol. Biol. 275, 3–16 (1998)
Fotjík, P., Vorlíckova, M.: The fragile X chromosome (GCC) repeat folds into a DNA tetraplex at neutral pH. Nucleic Acids Res. 29, 4684–4690 (2001)
Armentano, D., De Munno, G., Di Donna, L., Sindona, G., Giorgi, G., Salvini, L., Napoli, A.: Self-assembling of cytosine nucleoside into triply-bound dimers in acid media. A comprehensive evaluation of proton-bound pyrimidine nucleosides by electrospray tandem mass spectrometry, X-rays diffractometry, and theoretical calculations. J. Am. Soc. Mass Spectrom. 15, 268–279 (2004)
Day, H.A., Pavlou, P., Waller, Z.A.E.: i-Motif DNA: structures, stability, and targeting ligands. Bioinorg. Med. Chem. 22, 4407–4418 (2014)
Armentrout, P.B.: Mass spectrometry—not just a structural tool: the use of guided ion beam tandem mass spectrometry to determine thermochemistry. J. Am. Soc. Mass Spectrom. 13, 419–434 (2002)
Yang, B., Wu, R.R., Rodgers, M.T.: Base-pairing energies of proton-bound homodimers determined by guided ion beam tandem mass spectrometry: application to cytosine and 5-substituted cytosines. Anal. Chem. 85, 11000–11006 (2013)
Yang, B., Rodgers, M.T.: Base-pairing energies of proton-bound heterodimers of cytosine and modified cytosines: implications for the stability of DNA i-motif conformations. J. Am. Chem. Soc. 136, 282–290 (2014)
Yang, B., Moehlig, A.R., Frieler, C.E., Rodgers, M.T.: Base-pairing energies of protonated nucleobase pairs and proton affinities of 1-methylated cytosines: model systems for the effects of the sugar moiety on the stability of DNA i-motif conformations. J. Phys. Chem. B. 119, 1857–1868 (2015)
Yang, B., Wu, R.R., Rodgers, M.T.: Base-pairing energies of proton-bound dimers and proton affinities of 1-methyl-5-halocytosines: implications for the effects of halogenation on the stability of the DNA i-motif. J. Am. Soc. Mass Spectrom. 26, 1469–1482 (2015)
Yang, B., Rodgers, M.T.: Base-pairing energies of protonated nucleoside base pairs of dCyd and m5dCyd: implications for the stability of DNA i-motif conformations. J. Am. Soc. Mass Spectrom. 26, 1394–1403 (2015)
Rajabi, K., Theel, K., Gillis, E.A.L., Beran, G., Fridgen, T.D.: The structure of the protonated adenine dimer by infrared multiple photon dissociation spectroscopy and electronic structure calculations. J. Phys. Chem. A. 113, 8099–8107 (2009)
Cruz-Ortiz, A.F., Rossa, M., Berthias, F., Berdakin, M., Maitre, P., Pino, G.A.: Fingerprints of both Watson-Crick and Hoogsteen isomers of the isolated (cytosine-guanine)H+ pair. J. Phys. Chem. Lett. 8, 5501–5506 (2017)
Azargun, M., Fridgen, T.D.: Guanine tetrads: an IRMPD spectroscopy, energy resolved SORI-CID, and computational study of M(9-ethylguanine)4 + (M = Li, Na, K, Rb, Cs) in the gas phase. Phys. Chem. Chem. Phys. 17, 25778–257853 (2015)
Hoogsteen, K.: The crystal and molecular structure of a hydrogen-bonded complex between 1-methylthymine and 9-methyladenine. Acta Crystallogr. 16, 907–916 (1963)
Pramanik, S., Nakamura, K., Usui, K., Nakano, S.I., Saxena, S., Matsui, J., Miyoshi, D., Sugimoto, N.: Thermodynamic stability of Hoogsteen and Watson−Crick base pairs in the presence of histone H3-mimicking peptide. Chem. Comm. 47, 2790–2792 (2011)
Halder, A., Halder, S., Bhattacharyya, D., Mitra, A.: Feasibility of occurrence of different types of protonated base pairs in RNA: a quantum chemical study. Phys. Chem. Chem. Phys. 16, 18383–18396 (2014)
Nikolova, E.N., Kim, E., Wise, A.A., O’Brien, P.J., Andricioaei, I., Al-Hashimi, H.M.: Transient Hoogsteen base pairs in canonical duplex DNA. Nature. 470, 498–504 (2011)
Honig, B., Rohs, R.: Flipping Watson and Crick. Nature. 470, 472–473 (2011)
Alvey, H.S., Gottardo, F.L., Nikolova, E.N., Al-Hashimi, H.M.: Widespread transient Hoogsteen base pairs in canonical duplex DNA with variable energetics. Nat. Commun. 5, 4786/1–4786/8 (2014)
Povsic, T.J., Dervan, P.B.: Triple helix formation by oligonucleotides on DNA extended to the physiological pH range. J. Am. Chem. Soc. 111, 3059–3061 (1989)
Rajagopal, P., Feigon, J.: Triple-strand formation in the homopurine:homopyrimidine DNA oligonucleotides d(G-A)4 and d(T-C)4. Nature. 339, 637–640 (1989)
Mirkin, S.M.: Expandable DNA repeats and human disease. Nature. 447, 932–940 (2007)
Gacy, A.M., Goellner, G.M., Spiro, C., Chen, X., Gupta, G., Bradbury, E.M., Dyer, R.B., Mikesell, M.J., Yao, J.Z., Johnson, A.J., Richter, A., Melancon, S.B., McMurray, C.T.: GAA instability in Friedreich’s ataxia shares a common, DNA-directed and intraallelic mechanism with other trinucleotide diseases. Mol. Cell. 1, 583–593 (1998)
Jun, J., Han, S.Y.: Theoretical exploration of gas-phase conformers of proton-bound non-covalent heterodimers of guanine and cytosine rare tautomers: structures and energies. Theor. Chem. Accounts. 136, 136–146 (2017)
Han, S.Y., Lee, S.H., Chung, J., Oh, H.B.: Base-pair interactions in the gas-phase proton-bonded complexes of C+G and C+GC. J. Chem. Phys. 127, 245102–245110 (2007)
Park, J.J., Lee, C.S., Han, S.Y.: Proton transfer accounting for anomalous collision-induced dissociation of proton-bound Hoogsteen base pair of cytosine and guanine. J. Am. Soc. Mass Spectrom. 29, 2368–2379 (2018)
Hobza, P., Šponer, J.: Structure, energetics, and dynamics of the nucleic acid base pairs: nonempirical ab initio calculations. Chem. Rev. 99, 3247–3276 (1999)
Seong, Y., Han, S.Y., Jo, S.-C., Oh, H.B.: An anomalous dissociation of protonated cluster ions of DNA guanine-cytosine base-par. Mass Spectrom. Lett. 2, 72–75 (2011)
Cooks, R.G., Koskinen, J.T., Thomas, P.D.: The kinetic method of making thermochemical determinations. J. Mass Spectrom. 34, 85–92 (1999)
Cooks, R.G., Patrick, J.S., Kotiaho, T., McLuckey, S.A.: Thermochemical determinations by the kinetic method. Mass Spectrom. Rev. 13, 287–339 (1994)
Cheng, X., Wu, Z., Fenselau, C.: Collision energy dependence of proton-bound dimer dissociation: entropy effects, proton affinities, and intramolecular hydrogen-bonding in protonated peptides. J. Am. Chem. Soc. 115, 4844–4848 (1993)
Hunter, E.P.L., Lias, S.G.: Evaluated gas phase basicities and proton affinities of molecules: an update. J. Phys. Chem. Ref. Data. 27, 413–656 (1996)
Greco, F., Liguori, A., Sindona, G., Uccella, N.: Gas-phase proton affinity of deoxyribonucleosides and related nucleobases by fast atom bombardment tandem mass spectrometry. J. Am. Chem. Soc. 112, 9092–9096 (1990)
Kertesz, T.M., Hall, L.H., Hill, D.W., Granta, D.F.: CE50: quantifying collision induced dissociation energy for small molecule characterization and identification. J. Am. Soc. Mass Spectrom. 20, 1759–1767 (2009)
Frisch, M. J., Trucks, G. W., Schlegel, H. B., Scuseria, G. E., Robb, M. A., Cheeseman, J. R., Scalmani, G., Barone, V., Mennucci, B., Petersson, G. A., Nakatsuji, H., Caricato, M., Li, X., Hratchian, H. P., Izmaylov, A. F., Bloino, J., Zheng, G., Sonnenberg, J. L., Hada, M., Ehara, M., Toyota, K., Fukuda, R., Hasegawa, J., Ishida, M., Nakajima, T., Honda, Y., Kitao, O., Nakai, H., Vreven, T., Montgomery, Jr., J. A., Peralta, J. E., Ogliaro, F., Bearpark, M., Heyd, J. J., Brothers, E., Kudin, K. N., Staroverov, V. N., Keith, T., Kobayashi, R., Normand, J., Raghavachari, K., Rendell, A., Burant, J. C., Iyengar, S. S., Tomasi, J., Cossi, M., Rega, N., Millam, J. M., Klene, M., Knox, J. E., Cross, J. B., Bakken, V., Adamo, C., Jaramillo, J., Gomperts, R., Stratmann, R. E., Yazyev, O., Austin, A. J., Cammi, R., Pomelli, C., Ochterski, J. W., Martin, R. L., Morokuma, K., Zakrzewski, V. G., Voth, G. A., Salvador, P., Dannenberg, J. J., Dapprich, S., Daniels, A. D., Farkas, O., Foresman, J. B., Ortiz, J. V., Cioslowski, J., Fox, D. J.: Gaussian 09, Revision D.01, Gaussian, Inc., Wallingford CT (2013)
Boys, S.F., Bernardi, F.: The calculation of small molecular interactions by the differences of separate total energies. Some procedures with reduced errors. Mol. Phys. 100, 65–73 (2002)
Tomasi, J., Mennucci, B., Cammi, R.: Quantum mechanical continuum solvation models. Chem. Rev. 105, 2999–3094 (2005)
Peng, C., Schlegel, H.B.: Combining synchronous transit and quasi-Newton methods for finding transition states. Israel J. Chem. 33, 449–454 (1993)
Curtiss, L.A., Redfern, P.C., Raghavachari, K.: Gaussian-4 theory. J. Chem. Phys. 126, 084108 (2007)
Curtiss, L.A., Redfern, P.C., Raghavachari, K.: Gaussian-4 theory using reduced order perturbation theory. J. Chem. Phys. 127, 124105 (2007)
Montgomery Jr., J.A., Frisch, M.J., Ochterski, J.W., Petersson, G.A.: A complete basis set model chemistry. VI. Use of density functional geometries and frequencies. J. Chem. Phys. 110, 2822–2827 (1999)
Montgomery Jr., J.A., Frisch, M.J., Ochterski, J.W., Petersson, G.A.: A complete basis set model chemistry. VII. Use of the minimum population localization method. J. Chem. Phys. 112, 6532–6542 (2000)
Moser, A., Range, K., York, D.M.: Accurate proton affinity and gas-phase basicity values for molecules important in biocatalysis. J. Phys. Chem. B. 114, 13911–13921 (2010)
Kolboe, S.: Proton affinity calculations with high level methods. J. Chem. Theory Comput. 10, 3123–3128 (2014)
Somers, K.P., Simmie, J.M.: Benchmarking compound methods (CBS-QB3, CBS-APNO, G3, G4, W1BD) against the active thermochemical tables: formation enthalpies of radicals. J. Phys. Chem. A. 119, 8922–8933 (2015)
Ung, H.U., Huynh, K.T., Poutsma, J.C., Oomens, J., Berden, G., Morton, T.H.: Investigation of proton affinities and gas phase vibrational spectra of protonated nucleosides, deoxynucleosides, and their analogs. Int. J. Mass Spectrom. 378, 294–302 (2015)
Li, A., Muddana, H.S., Gilson, M.K.: Quantum mechanical calculation of noncovalent interactions: a large-scale evaluation of PMx, DFT, and SAPT approaches. J. Chem. Theory Comput. 10, 1563–1575 (2014)
Burns, L.A., Vázquez-Mayagoitia, Á., Sumpter, B.G., Sherrill, C.D.: Density-functional approaches to noncovalent interactions: a comparison of dispersion corrections (DFT-D), exchange-hole dipole moment (XDM) theory, and specialized functionals. J. Chem. Phys. 134, 084107 (2011)
Acknowledgements
This work was supported by MSIT via NRF (Grant No. NRF-2016R1D1A1B03931987). This work was also supported by MOTIE via KEIT (Grant No. 10063335).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
ESM 1
(PDF 696 kb)
Rights and permissions
About this article
Cite this article
Park, J.J., Han, S.Y. Alternated Branching Ratios by Anomaly in Collision-Induced Dissociation of Proton-Bound Hoogsteen Base Pairs of 1-Methylcytosine with 1-Methylguanine and 9-Methylguanine. J. Am. Soc. Mass Spectrom. 30, 846–854 (2019). https://doi.org/10.1007/s13361-019-02161-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-019-02161-2