Abstract
Sulfotyrosine and phosphotyrosine are two post-translational modifications present in higher eukaryotes. A simple and direct mass spectrometry method to distinguish between these modifications is crucial to advance our understanding of the sulfoproteome. While sulfation and phosphorylation are nominally isobaric, the accurate mass of the sulfuryl moiety is 9.6 mDa less than the phosphoryl moiety. Based on this difference, we have used an Orbitrap Fusion Lumos mass spectrometer to characterize, resolve, and distinguish between sulfotyrosine and phosphotyrosine modifications using a set of model peptides. Multiple fragmentation techniques, namely HCD, CID, ETD, ETciD, and EThcD, have been used to compare the different fragmentation behaviors between peptides modified with these species. Sulfotyrosine undergoes neutral loss using HCD and CID, but the sulfuryl moiety is largely stable under ETD. In contrast, phosphotyrosine is stable during fragmentation using all these methods. This differential stability provides a mechanism to distinguish sulfopeptides from phosphopeptides. Based on the rigorous characterization presented herein, this work serves as a model for accurate identification of phosphotyrosine and, more challenging, sulfotyrosine, in complex proteomic samples.

ᅟ
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
Sulfotyrosine and phosphotyrosine both exist in higher eukaryotes. Unlike phosphorylation, which has been extensively studied [1,2,3], much less is known about the extent of sulfation within a proteome. Due to its biological importance in processes as varied as viral infection, immune regulation, and hemostasis, sulfation has become the focus of more intensive research efforts [4, 5]. The sulfation reaction is mediated by two enzymes, tyrosylprotein sulfotransferase 1 or 2 (TPSTs) in humans. As these enzymes are Golgi-resident, sulfation is added only to membrane and secreted proteins (i.e., proteins that transit through the secretory pathway) [5, 6]. Previous pulse-chase experiments suggest that approximately 3% of the tyrosine residues on secreted proteins are sulfated [7, 8]. To date, only 48 human sulfoproteins have been annotated in Uniprot, suggesting that a large fraction of the sulfoproteome remains to be identified. Identification of sulfotyrosine modified proteins in a complex proteome is technically challenging for multiple reasons. First, sulfoproteins have been traditionally identified using radiolabeling [9,10,11,12,13,14,15,16]. This is by far the most reliable method to distinguish sulfation from phosphorylation in biological samples; however, this method is labor intensive, requires significant starting material, and does not interface well with mass spectrometry. Second, the nominally isobaric nature of sulfation and phosphorylation modifications poses a challenge for mass spectrometric approaches. Multiple mass spectrometry methods have been applied to distinguish these two modifications [17,18,19]. However, work to date has focused on individual peptides and/or MS-level differences in synthetic standards, and the applicability of the current findings for large-scale sulfoproteome analysis remains unclear [17,18,19]. Third, in a similar manner to phosphoserine and phosphothreonine, sulfotyrosine is highly labile during collision induced dissociation (CID), limiting our ability to localize this modification within a peptide sequence [20,21,22]. This lability is particularly problematic because unlike phosphoserine and phosphothreonine, which undergo beta elimination, the sulfryl group is lost in a neutral fashion, making the exact site of modification on the peptide impossible to determine when more than one possibly modified residue exists.
As part of our ongoing efforts to understand the prevalence of sulfation in the human proteome, we have provided a framework to address these limitations. Specifically, we have used an Orbitrap Fusion Lumos with a high field detector to examine eight sets of synthetic peptides that have been modified with sulfate and phosphate. To our knowledge, this is the first time that a commercially available mass spectrometer has been used to resolve phosphopeptides from their sulfate-modified counterparts based upon their mass difference. In addition, we provide a systematic comparison on fragmentation techniques (specifically HCD, CID, ETciD, and EThcD) for the analysis of sulfopeptides versus phosphopeptides.
Materials and Methods
Peptide Synthesis
Sulfopeptides and phosphopeptides were synthesized using Fmoc solid-phase synthesis as previously described [23]. Fmoc-Tyr(SO3nP)-OH and Fmoc-Tyr(PO(OBzl)OH)-OH were purchased from Merck Millipore (Burlington, MA, USA). Sulfopeptides were deprotected in 2 M ammonium acetate at 37 °C before use.
ESI-MS/MS
ESI-MS was carried out using an Orbitrap Fusion Lumos (Thermo Fisher Scientific, San Jose, CA, USA). Peptides were directly infused using a syringe pump with an infusion rate of 10 uL/min. For the accurate mass comparison experiment, nominally isobaric sulfo- and phosphopeptides were mixed at approximately equal concentrations to yield equivalent signal intensities in the instrument. The accurate mass was measured using resolutions of 240,000 and 500,000 FWHM with a total of 20 scans acquired. CID and HCD spectrum were collected at 35% and 25% energy, respectively. Electron transfer dissociation (ETD) reaction time was set to 50 ms in all experiments. The supplemental activation energy in ETciD and EThcD varied from 10% to 45%.
Identification of Sulfopeptides Spiked into a Serum Digest
Eight sulfopeptides were mixed with trypsin-digested serum in 1:200, 1:1200, and 1:2000 (w/w) ratios (5, 0.83, and 0.5 ng sulfopeptide in 1 ug serum) and analyzed by LC-MS/MS with multistage activation. Peptides were separated on an Acclaim PepMap RSLC C18 analytical column (75 μm × 150 mm, 2 μm, 100 Å). The elution gradient was as follows: 0–0.5 min, 2%–6% B; 0.5–105.5 min, 6%–40% B; 105.5–115 min, 40%–100% B; 115–120 min, 100% B, followed by re-equilibration to 2% B (Solvent A: 0% acetonitrile, 0.1% formic acid; Solvent B: 80% acetonitrile, 0.1% formic acid). The mass spectrometer was run with (and without) an inclusion list to specifically sequence masses corresponding to the eight sulfopeptides. Identification of a given sulfopeptide was done by ProteinProspector ver. 5.20.0 searching against a database that included all Homo sapien proteins in the Uniprot database (downloaded 2016-9-6, 154,578 entries), to which our eight sequences had been appended.
Result and Discussion
High Resolution Mass Spectra
We synthesized a set of eight sulfopeptides using sites that have been determined previously to be sulfated in humans (Table 1) [24,25,26,27,28,29,30]. For each of these peptides, we also synthesized a second set wherein the sulfotyrosine was replaced with phosphotyrosine. In all cases, we synthesized 11-mer peptides with the modified residue in the central position. Since the sequences are identical, and the modifications are nominally isobaric, these eight peptide pairs have the same nominal mass. However, the exact monoisotopic mass of sulfate is 9.6 mDa less than for phosphate. The 9.6 mDa change in mass results from the nuclear binding energy difference between the two, despite the fact that 31P+H and 32S both have the same number of neutrons and protons. We took advantage of this very minor difference to distinguish these two modifications. Tryptic peptides usually show up as charge state z = 2+, which creates a 4.8 mDa Thomson difference between them. Specialized mass spectrometers, e.g., Fourier transform ion cyclotron resonance (FT-ICR) instruments with mass revolving power of 10 [6], are capable of resolving this difference in sulfated threonine [22, 31]. We asked whether the Orbitrap Fusion Lumos design for high-throughput proteomics can also distinguish this difference in practice [32,33,34]. Thus, we mixed the sulfated and phosphorylated peptides at a 1:1 ratio and analyzed spectra collected at two resolutions (240,000 and 500,000) to identify the minimum resolution required to resolve isotope peaks. Two specific examples are shown in Figure 1. The peptides SSGADs/pYPDELQ (TRY1) and ISDRDs/pYMGWMD (SCG2) were chosen because they are the lowest and highest mass among our eight peptides, respectively. The theoretical m/z values for a charge state of two are 630.7379 and 630.7426, 748.7507 and 748.7507, respectively. Using 240,000 resolution, we were able to separate the TRY1 peptides into two distinct peaks (Figure 1a). However, at that resolution, we were unable to do so for SCG2 peptide. Nevertheless, the width of that single peak was sufficiently broad to indicate that it contained more than one peptide (Figure 1b). Increasing the resolution to 500,000 gives better separation for TRY1, with sharper peaks. Sulfo- and phospho-SCG2 are well resolved using this resolution. In summary, this experiment demonstrates that we are capable of using accurate mass to distinguish species that vary in mass by as little as 9.6 mDa. In general, a resolution of 240,000 is sufficient to resolve peptides different by 9.6 mDa when the overall mass of the peptides is around 1500 Da or below (see Supplementary Figure S1). We should note that it is significantly easier to separate a phosphopeptide from its sulfopeptide analog in the MS than it is to assign an unknown peptide as phosphorylated or sulfated. This is due to the fact that the mass of the measured peptide may fall in between that of the phosphopeptide and sulfopeptide, thus effectively doubling the required mass accuracy search window.
Resolving sulfated and phosphorylated peptides by 240,000 and 500,000 resolution power. Sulfated and phosphorylated TRY1 and SCG2 have monoisotopic masses of (630.7379) and (630.7426); (748.7507) and (748.7554), respectively. Additional peptide data are provided in Supporting Material
Our work represents the first case in which sulfation and phosphorylation modifications are distinguished solely by the exact m/z with a commercial mass spectrometer. We should note that the 240,000 resolution scans to a total of 0.6 s, whereas the scans at 500,000 took 1.2 s. This will necessarily affect the cycle time during a large-scale experiment aimed at identifying novel sulfopeptides.
Comparing CID, HCD, and ETD Spectra of Sulfated or Phosphorylated Peptides
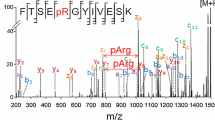
We then investigated the degree to which HCD and CID can distinguish sulfation from phosphorylation using our model peptides. All samples were run using these methods, and data from peptides ISDRDs/pYMGWMD are shown in Figure 2. HCD fragmentation results in a large number of fragment ions for this sulfopeptide; however, none of these fragment ions were observed retaining the sulfuryl (SO3) moiety (Figure 2a). In contrast, fragment ions from the phosphotyrosine peptide retained the phosphoryl group on the tyrosine (Figure 2b). We then compared these spectra with those obtained by CID. Previous studies showed that during CID, sulfotyrosine undergoes neutral loss of 80 Da whereas phosphotyrosine does not [20, 35]. These experiments were either done in negative ion mode [35] or using sulfo- and phosphopeptides that did not share the same sequence [20]. In addition, CID has been combined with ETD to characterize synthetic sulfothreonine modified peptides [31]. As a prelude for a large-scale sulfotyrosine proteomic characterization, we sought to better define how sulfotyrosine-modified peptides behaved in an Orbitrap Fusion Lumos, an instrument that has significant design differences from previous generations. All eight of our sulfopeptides experienced significant neutral loss during CID (see Figure 2c and Supplementary Figure S2). For each spectrum, the neutral loss peak [M-80]2+ represented between 27% and 95% of the total ion current. This wide range is due to the fact that the neutral loss peak often undergoes further loss of ammonia/water (see Supplementary Figures S2–4C). In contrast, phosphopeptides undergo fragmentation with the phosphoryl moiety intact (Figure 2d). In these cases, we only observed a neutral loss peak corresponding to 0.2% or less of the total ion current. These results largely agree with the previous studies. We took this a step further to see whether we can recover sequence information from the sulfopeptide CID spectra. Figure 2c shows a zoomed-in view of fragment ion intensity of the sulfopeptide spectra. We can clearly find fragment ions corresponding to the y-, b-, y-80-, and b-80-ion series. Although the relative signal is weak, we were able to assign the fragment ions, an advance not previously reported. Ion trap-style instruments are also capable of obtaining CID spectra using multistage activation. During this acquisition approach, the precursor ion is vibrationally excited at the same time as any predefined neutral loss fragments. Multistage activation produced MS/MS spectra containing large numbers of b- and y-ions, sufficient to allow database searching; however, none of these product ions retained the sulfuryl moiety (Figure 2e). The labile nature of sulfate during CID and HCD provides a way to unambiguously distinguish between these two modifications during the peptide identification process, providing that a subset of the peaks matched during MS/MS span the modification site.
HCD, CID, and multistage-activation CID spectra of sulfated and phosphorylated CCKN. (a) HCD spectrum of sulfopeptide. (b) HCD spectrum of phosphopeptide. (c) CID spectrum of sulfopeptide. (d) CID spectrum of phosphopeptide. (e) Multistage activation CID spectrum of sulfopeptide. Additional peptide data are provided in Supporting Material
To address the issue of labile neutral loss, we examined how sulfotyrosine-containing peptides behaved during ETD. ETD is known to provide peptide backbone cleavage while largely preserving labile side chain modifications [36]. It has also been applied to the study of glycopeptides, allowing determination of glycopeptide sequences [37]. Adding supplemental activation such as CID and HCD increases the efficiency of fragment ion generation. These hybrid approaches combining ETD and CID or HCD are termed ETciD and EThcD, respectively [38, 39]. We examined the fragmentation behavior using these approaches for our entire peptide panel using 35% supplemental energy (Figure 3 and Supplementary Figure S3). We should note that all of our synthetic peptides assumed a charge state of plus two during electrospray. While the majority of peptides generated in a tryptic digest will have a charge state of plus two, ETD-style approaches have been shown to work best with charge states of three and higher [40]. In Figure 3, we present ETD, ETciD, and EThcD spectra of ISDRDs/pYMGWMD. ETD, and ETciD fragments of phosphorylated peptides all retain their modifications. In contrast, mild neutral loss is still observed in sulfated peptides (Figure 3 and Supplementary Figure S3: A, B, C, and D show the baseline region). EThcD generated more fragments for both sulfated and phosphorylated peptides. However, c-/z- ions of sulfopeptides underwent significant neutral loss at this HCD energy whereas their phosphorylated counterparts did not (Figure 3e and f). Noticeably, EThcD generated significant levels of y- and b-ions, though none of these fragments retained sulfation (spectra for the additional seven peptides are shown in Supplementary Figure S3). In contrast to HCD or CID fragmentation alone, our main interest in using ETD is the generation of fragments bearing the sulfation to enable site localization of this PTM. Therefore, we then examined the supplemental energy that maximizes the intensity of c- and z-ions.
ETD, ETciD, and EThcD spectra of sulfated and phosphorylated CCKN. (a) ETD spectrum of sulfopeptide. (b) ETD spectrum of phosphopeptide. (c) ETciD spectrum of sulfopeptide. (d) ETciD spectrum of phosphopeptide. (e) EThcD spectrum of sulfopeptide. (f) EThcD spectrum of phosphopeptide. Additional peptide data are provided in Supporting Material
Fragmentation Efficiency as a Function of Supplemental Energy
The ETciD and EThcD shown in Figure 3 were run at a supplemental energy of 35%. To obtain a better understanding of how supplemental energy affected ETD, we conducted ETciD and EThcD at energies from 10% to 45% using all 16 peptides with a fixed ETD reaction time of 50 ms, calculating the relative abundance of selected sulfation/phosphorylation-containing peaks normalized to total ion current (Figure 4). For the majority of the 16 sulfated and phosphorylated peptides, none of the ETciD energies tested led to widespread increases in c- and z-ion abundance. Of the 79 c- and z-ions, the intensities of which we determined as a function of supplemental energy, for only 6 ions (7.6%) did ETciD lead to an increase in abundance of 25% or more relative to ETD alone (Figure 4a and b). The peptides used in this study were all doubly charged, and doubly charged peptides are known to undergo less effective ETD fragmentation than those with higher charge states. However, previous studies have demonstrated that ETciD give clear improvement for doubly charged peptides [39]. To determine if the lack of improvement was due to the fact that we were analyzing modified peptides, we examined the behavior of angiotensin I (DRVYIHPFHL) during ETciD. In the case of the triply charged angiotensin I precursor, we observed that many c- and z-ions increased in abundance approximately 50% when fragmenting using ETciD relative to ETD alone. However, ETciD of the doubly charged precursor yielded spectra that were largely similar to those generated with ETD alone (Supplementary Figure S4). While ETciD of doubly charged peptides has previously been shown to be beneficial, those experiments were conducted in an LTQ, where the ETD reaction and CID activation were applied in the same compartment. In contrast, in our Orbitrap Fusion Lumos, ETD occurs in the ion routing multipole, and peptides are subsequently transferred to the ion trap for supplemental CID. We suspect that this spatial and temporal separation of ETD and CID limits the ability of CID to promote ETD-type fragmentation (at least for doubly charged peptides).
The percentage of major c-/z- ions from all 16 peptides using ETciD and EThcD energy: 0, 10, 15, 20, 25, 30, 35, 40, and 45%. (a) The percentage of c-/z- ions of sulfopeptides in ETciD. (b) The percentage of c-/z- ions of phosphopeptides in ETciD. (c) The percentage of c-/z- ions of sulfopeptides in EThcD. (d) The percentage of c-/z- ions of phosphopeptides in EThcD. (e) The percentage of c-80/z-80 ions of sulfopeptides in EThcD
In the EThcD experiment, the abundance of sulfation-containing c- and z-ions decreases with increasing energy (Figure 4c). To determine whether this decrease was due to neutral loss of sulfuryl group from these ions, we monitored for the presence of desulfated ions (c-80 and z-80). As shown in Figure 4e, supplemental HCD results in desulfation of these ions. Even at supplemental HCD energies of 10%–15%, which are more typically used, we observed loss of sulfuryl group from c- and z-ions. For none of EThcD-generated c- and z-ions examined we observe an increase in ion intensity with any level of supplemental HCD energy. In contrast, the phosphopeptide fragmentation is improved by supplemental HCD with phosphoryl moiety remaining intact and c- and z-ions bearing this modification reaching a maximum at ~35% energy (Figure 4d). Altogether, this analysis suggests that ETD and/or ETciD are superior to EThcD for the analysis of sulfopeptides with respect to retention of the modification.
Recovery of Sulfopeptides from Complex Background
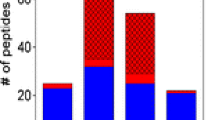
To test whether sulfopeptides can be identified in complex samples, we mixed sulfopeptide standards with trypsin-digested human serum at ratios of 1:200, 1:1200, and 1:2000 (w/w). Using a targeted MS/MS acquisition approach with an inclusion list, the resulting database search (i.e., Uniprot Homo sapiens plus our eight peptides) from the 1:200 mixture identified all eight peptides. Further increasing the ratio of sulfopeptides to serum to 1:1200 resulted in only six being successfully identified, and increase in ratio to 1:2000 resulted in identification of only three sulfopeptides. In a parallel experiment in which our sulfopeptides were not specifically targeted for fragmentation, we saw a reduction in the number of sulfopeptides identified to six, two, and two, respectively, from the 1:200, 1:1200, and 1:2000 mass ratio spiked samples. Figure 5 shows the base peak chromatogram of the 1:2000 dilution for our targeted experiment with the locations of the sulfopeptide ions marked by arrows. The sulfopeptide standards eluted with a broad range of retention times. The probability of generating interpretable MS2 spectra depends on the complexity of the sample as well as the ionization efficiency of the peptides in question. Supplementary Figure S5 shows the MS1 spectra of sulfopeptides from the 1:1200 dilution sample. The ionization efficiencies of our sulfopeptide standards are fairly low. The low ionization efficiency is likely due to the fact these peptides do not contain an arginine or lysine residue at their carboxyl termini (although those peptides from CXCR4 and CCKN contain internal lysine and arginine residues, respectively). In addition, the negatively charged sulfuryl groups on these peptides likely decrease their ionizability and/or lower their charge state. In order to identify the majority of our sulfopeptide standards spiked into one microgram of serum digest, we required approximately 0.83 ng of each peptide. However, the limit of detection for fully tryptic sulfopeptides can reasonably be expected to be much lower than our non-tryptic sulfopeptide standards. The low abundance and low ionization efficiency of sulfopeptides indicate that large-scale characterization of the sulfoproteome will require targeted sulfopeptide enrichment.
The base peak chromatogram resulting from sulfopeptides spiked into a human serum digest at 1:2000 mixture with MS/MS acquisition collected using a data-dependent acquisition approach, which incorporated an inclusion list for the our sulfopeptide standards. While none of the sulfopeptides were the base peak, the arrows indicate their elution positions based upon having been identified in 1:200 ratio. Upper right corner: the number of sulfopeptides identified from the 1:200, 1:1200, and 1:2000 ratio dilution experiments (asterisks indicate sulfopeptides identified in a parallel experiment, which lacked an inclusion list). To note: the peptide from protein TRY1 and F81 was identified from the database search in the 1:2000 dilution but not the 1:1200 dilution. The MS/MS for this peptide in the 1:1200 dilution was presumably of low quality, and a better one was not also obtained due to the dynamic exclusion window
Conclusion
This work evaluates multiple mass spectrometric approaches for distinguishing between sulfated and phosphorylated peptides. The mass spectrometer used in this study has a maximum resolving power of 500,000 FWHM. At this resolving power, we were able to distinguish the 9.6 mDa difference between sulfo- and phosphopeptides for peptides with masses up to 1.5 kDa. With respect to fragmentation, sulfation and phosphorylation displayed distinct fragmentation patterns in CID and HCD, with phosphate being retained on the tyrosine residue, whereas the sulfuryl moiety is lost. Both multistage activation CID and HCD yielded fragmentation with significant numbers of y- and b-type ions, enabling identification of the peptide sequence, but the resulting neutral loss of sulfate would make it impossible to determine the site of sulfation in cases where more than one potential site existed. For site localization, it is necessary to employ ETD or ETciD, which are both able to preserve sulfation site information. Owing to the highly labile nature of sulfation, EThcD causes loss of sulfate, even at moderately low energies. Based on these results as a whole, we would recommend that studies aimed at characterizing the sulfo-proteome should adopt a two pronged approach, where either multistage activation CID or HCD is used to first sequence unknown peptides. ETciD should then be used to provide any additional information necessary to precisely localize the site of modification. Finally, our data show that untargeted identification of sulfopeptides from a complex digest likely would require enrichment, or at least fractionation, of the peptide mixture. This would be necessary to decrease complexity and allow for identification of sulfopeptides, which would otherwise not be selected for sequencing due to their low abundances and poor ionization properties.
References
Trinidad, J.C., Specht, C.G., Thalhammer, A., Schoepfer, R., Burlingame, A.L.: Comprehensive identification of phosphorylation sites in postsynaptic density preparations. Am. Soc. Biochem. Mol. Biol. 5.5, 914–922 (2006)
Judit Ville, S.A.B., Gerber, S.A., Gygi, S.P.: Large scale phosphorylation analysis of mouse liver. Nat. Acad. Sci. USA. 104, 1488–1493 (2007)
Bo Zhai, J.V., Beausoleil, S.A., Mintseris, J., Gygi, S.P.: Phosphoproteome analysis of Drosophila melanogaster embryos. J. Proteome Res. 7, 1675–1682 (2007)
Yang, Y.S.: Tyrosine sulfation as a protein post-translational modification. Molecules. 20, 2138–2164 (2015)
Monigatti, F., Hekking, B., Steen, H.: Protein sulfation analysis – a primer. Biochim. Biophys. Acta. 176, 1904–1913 (2006)
Moore, K.L.: The biology and enzymology of protein tyrosine O-sulfation. J. Biol. Chem. 278(27), 24243–24246 (2003)
Annette Hille, T.B., von Figura, K., Huttner, W.B.: Occurrence of tyrosine sulfate in proteins secretory and lysosomal a balance sheet secretory and lysosomal proteins. Eur. J. Biochem. 188, 577–586 (1990)
Hille, A., Huttner, W.B.: Occurrence of tyrosine sulfate in proteins membrane proteins membrane proteins. Eur. J. Biochem. 188, 587–596 (1990)
Yun, H.Y., Keutmann, H.T., Eippers, B.A.: Alternative splicing governs sulfation of tyrosine or oligosaccharide on peptidylglycine a-amidating monooxygenase. J. Biol. Chem. 269, 10946–10955 (1994)
Preobrazhensky, A.A., Dragan, S., Kawano, T., Gavrilin, M.A., Gulina, I.V., Chakravarty, L., Kolattukudy, P.E.: Monocyte chemotactic protein-1 receptor CCR2B is a glycoprotein that has tyrosine sulfation in a conserved extracellular N-terminal region. J. Immunol. 165, 5295–5303 (2000)
Michael Farzan, T.M., Kolchinsky, P., Wyatt, R., Mark Cayabyab, N.P.G., Gerard, C., Sodroski, J., Choe, H.: Tyrosine sulfation of the amino terminus of CCR5 facilitates HIV-1 entry. Cell 96, 667–676 (1999)
Michael Farzan, C.E.S., Vasilieva, N., Leung, D., Kuhn, J., Gerard, C., Gerard, N.P., Choe, H.: Sulfated tyrosines contribute to the formation of the C5a docking site of the human C5a anaphylatoxin receptor. J. Exp. Med. 193, 1059–1065 (2001)
Forbes, E.G., Cronshaw, A.D., MacBeath, J.R.E., Hulmes, D.J.S.: Tyrosine-rich acidic matrix protein (TRAMP) is a tyrosine-sulphated and widely distributed protein of the extracellular matrix. FEBS Lett. 351, 433–436 (1994)
Gao, J., Choe, H., Bota, D., Wright, P.L., Gerard, C., Gerard, N.P.: Sulfation of tyrosine 174 in the human C3a receptor is essential for binding of C3a anaphylatoxin. J. Biol. Chem. 278, 37902–37908 (2003)
Benedum, U.M., Lamouroux, A., Konecki, D.S., Rosa, P., Hille, A., Baeuerle, P.A., Frank, R., Lottspeich, F., Mallet, J., Huttner, W.B.: The primary structure of human secretogranin I chromogranin B.comparison with chromogranin A reveals homologous terminal domains and a large intervening variable region. EMBO J.. 6, 1203–1211 (1987)
Hortin, G., Fok, K.F., Toren, P.C., Strauss, A.W.: Sulfation of a tyrosine residue in the plasmin-binding domainof a2-antiplasmin. J. Biol. Chem. 262, 3082–3085 (1987)
Patrick, A.L., Stedwell, C.N., Polfer, N.C.: Differentiating sulfopeptide and phosphopeptide ions via resonant infrared photodissociation. Anal. Chem. 86, 5547–5552 (2014)
Hersberger, K.E., Hakansson, K.: Characterization of O-sulfopeptides by negative ion mode tandem mass spectrometry: superior performance of negative ion electron capture dissociation. Anal. Chem. 84, 6370–6377 (2012)
Drake, S.K., Hortin, G.L.: Improved detection of intact tyrosine sulfate-containing peptides by matrix-assisted laser desorption/ionization time-of-flight mass spectrometry in linear negative ion mode. Int. J. Biochem. Cell Biol. 42, 174–179 (2010)
Nemeth-Cawley, J.F., Karnik, S., Rouse, J.C.: Analysis of sulfated peptides using positive electrospray ionization tandem mass spectrometry. J. Mass Spectrom. 36, 1301–1311 (2001)
Yagami, T., Kitagawa, K., Aida, C., Fujiwara, H., Futaki, S.: Stabilization of a tyrosine o sulfate residue by a cationic functional group: formation of a conjugate acid base pair. J. Pept. Res. 56, 239–249 (2000)
Senko, M.W., Hendrickson, C.L., Pasa-Tolic, L., Marto, J.A., White, F.M., Guan, S., Marshall, A.G.: Electrospray ionization Fourier transform ion cyclotron resonance at 9.4 t. Rapid Commun. Mass Spectrom. 10, 1824–1828 (1996)
Simpson, L.S.: Regulation of chemokine recognition by site-specific tyrosine sulfation of receptor peptides. Chem. Biol. 16, 153–161 (2009)
Sahin-Toth, M., Kukor, Z., Nemoda, Z.: Human cationic trypsinogen is sulfated on Tyr154. FEBS J. 273, 5044–5050 (2006)
Vishnuvardhan, D., Beinfeld, M.C.: Role of tyrosine sulfation and serine phosphorylation in the processing of procholecystokinin to amidated cholecystokinin and its secretion in transfected AtT-20 Cells. Biochemistry. 39, 13825–13830 (2000)
Farzan, M.: The role of post-translational modifications of the CXCR4 amino terminus in stromal-derived factor 1 alpha association and HIV-1 entry. J. Biol. Chem. 277, 29484–29489 (2002)
Pittman, D.D., Wang, J.H., Kaufman, R.J.: Identification and functional importance of tyrosine sulfate residues within recombinant factor VI11. Biochemistry. 31, 3315–3325 (1991)
Arruda, V.R., Hagstrom, J.N., Deitch, J., Heiman-Patterson, T., Camire, R.M., Chu, K., Fields, P.A., Herzog, R.W., Couto, L.B., Larson, P.J., Katherine, A.: High, post-translational modifications of recombinant myotube synthesized human factor IX. Blood 97, 130–137 (2000)
Gerdes, H.-H., Rosa, P., Phillips, E., Baeuerle, P.A., Frank, R., Argos, P., Huttner, W.B.: The primary structure of human secretogranin II a widespread tyrosine-sulfated secretory granul Pe rotein that exhibits low pH- and calcium-induced aggregation. J. Biol. Chem. 264, 12009–12015 (1989)
Benjannet, S.: The proprotein convertase (PC) PCSK9 is inactivated by furin and/or PC5/6A: functional consequences of natural mutations and post-translational modifications. J. Biol. Chem. 281, 0561–0572 (2006)
Pattky, M.: Structure characterization of unexpected covalent O-sulfonation and ion-pairing on an extremely hydrophilic peptide with CE-MS and FT-ICR-MS. Anal. Bioanal. Chem. 407(22), 6637–6655 (2015)
Medzihradszky, K.F.: Sulfopeptide fragmentation in electron-capture and electron-transfer dissociation. J. Am. Soc. Mass Spectrom. 18(9), 1617–1624 (2007)
Hebert, A.S., Richards, A.L., Bailey, D.J., Ulbrich, A., Coughlin, E.E., Westphall, M.S., Coon, J.J.: The one hour yeast proteome. Mol. Cell. Proteomics. 13.1, 339–347 (2013)
Espadas, G., Borràs, E., Chiva, C., Sabidó, E.: Evaluation of different peptide fragmentation types and mass analyzers in data-dependent methods using an Orbitrap Fusion Lumos Tribrid mass spectrometer. Proteomics. 17(9), 1–9 (2017)
Edelson-Averbukh, M.: Discrimination between peptide O-sulfo- and O-phosphotyrosine residues by negative ion mode electrospray tandem mass spectrometry. J. Am. Soc. Mass Spectrom. 22, 2256–2268 (2011)
Zubarev, R.A.: Reactions of polypeptide ions with electrons in the gas phase. Mass Spectrom. Rev. 22, 57–77 (2003)
Trinidad, J.C., Schoepfer, R., Burlingame, A.L., Medzihradszky, K.F.: N- and O- glycosylation in the murine synaptosome. Mol. Cell. Proteomics. 12(12), 3474–3488 (2013)
Frese, C.K.: Toward full peptide sequence coverage by dual fragmentation combining electron-transfer and higher-energy collision dissociation tandem mass spectrometry. Anal. Chem. 84(22), 9668–9673 (2012)
Swaney, D.L., McAlister, G.C., Wirtala, M., Schwartz, J.C., Coon, J.J., Syka, J.E.P.: Supplemental activation method for high efficiency electron-transfer dissociation of doubly protonated peptide precursors. Anal. Chem. 79, 477–485 (2007)
Pitteri, S.J., Chrisman, P.A., Hogan, J.M., McLuckey, S.A.: Electron transfer ion/ion reactions in a three-dimensional quadrupole ion trap: reactions of doubly and triply protonated peptides with SO2. Anal. Chem. 77, 1831–1839 (2005)
Acknowledgments
This work was supported by the Indiana University College of Arts and Sciences Faculty Research Support SEED grant. The authors acknowledge David Smiley of the DiMarchi research group for his assistance with peptide synthesis.
Author information
Authors and Affiliations
Corresponding authors
Electronic Supplementary Material
ESM 1
(PDF 2499 kb)
Rights and permissions
About this article
Cite this article
Chen, G., Zhang, Y., Trinidad, J.C. et al. Distinguishing Sulfotyrosine Containing Peptides from their Phosphotyrosine Counterparts Using Mass Spectrometry. J. Am. Soc. Mass Spectrom. 29, 455–462 (2018). https://doi.org/10.1007/s13361-017-1854-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-017-1854-1