Abstract
Matrix assisted laser desorption ionization imaging mass spectrometry (MALDI-IMS) is a technique that has seen a sharp rise in both use and development. Despite this rapid adoption, there have been few thorough investigations into the actual physical mechanisms that underlie the acquisition of IMS images. We therefore set out to characterize the effect of IMS laser ablation patterns on the surface of a sample. We also concluded that the governing factors that control spatial resolution have not been correctly defined and therefore propose a new definition of resolution.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Matrix assisted laser desorption ionization mass spectrometry (MALDI-MS) is a technology that has been in use since the late 1980s in a wide range of applications from the analyses of bacterial colonies [1] and purified biomolecules [2] to the imaging of tissue sections [3]. MALDI is a technique whereby molecules are irradiated with a UV laser that, with the help of a co-crystallized organic matrix, causes the generation of an ion plume [4]. The resulting ions are then measured most often by time of flight mass spectrometry (TOF-MS). One of the more atypical uses for MALDI MS is the imaging of tissue sections (IMS) to show the location and relative intensity of individual biomolecules in the form of a spatial map. This technique is a methodological deviation from traditional MALDI MS analysis and, as such, requires highly specialized sample preparation and custom analysis methods, i.e., samples must be coated with matrix in a way that prevents delocalization of the analyte and the ablation pattern must be strictly controlled to generate pixels in specific X,Y coordinates rather than randomly rastering across the surface of the sample [5]. While performing our own IMS-based investigations [5, 6], we raised the question of what effect the constant laser bombardment was having, either mechanically or chemically, on the sample, as it was being ablated in the specific pattern required for IMS. At the time of writing, no other investigations of this nature had been performed. Therefore, we set out to thoroughly investigate and characterize the effect of grid laser ablation on samples during IMS analysis.
Experimental
Slide Preparation
In order to effectively visualize the ablation pattern of the laser, sample slides were prepared without any form of biological sample and simply consisted of specially prepared liquid nitrocellulose-coated indium tin oxide glass slides that were coated with MALDI matrix.
Slides were prepared in a similar fashion to previously published methodologies [5]. In short, standard sized indium tin oxide (ITO; LaserBio Labs, Valbonne, France) coated microscope slides were cut in half and coated with a thin film of liquid nitrocellulose (NC). The slide is then weighed on an analytical balance capable of accuracy to 5 decimal places and then affixed to the inside of the cooling finger of a modified sublimation apparatus as described previously [5].
Three hundred mg of sinapinic acid matrix was placed in the bottom petri dish and the chamber sealed. The cooling finger was packed with ice and the chamber evacuated to 25 mTorr. Sublimation was then performed in an oil bath at 200–210 °C for 45 min to achieve an ideal matrix coverage of 0.2 mg/cm2.
Mass Spectrometric Analysis
Following sublimation, the samples were imaged with a 5800 MALDI TOF/TOF (AB Sciex, Framingham USA) using a 355 nm Nd YAG laser via the TOFTOF Imaging software suite (AB Sciex, Framingham USA) in positive ion reflector mode with the following settings: laser power is set to 5100, digitizer vertical scale set to 0.05, and bandwidth at 20 MHz, mass range set to 3000–20,000 Da. Lateral resolution was set to 60 μm and laser shots were set to 200 per spot.
Scanning Electron Microscopy Instrumentation and Data processing
Once ablated, sample slides were then prepared for scanning electron microscopy. Sample slides were mounted onto adhesive carbon specimen stubs via the application of silver paint (ProSciTech). A MED 010 Balzers union carbon coater was used to vacuum deposit carbon onto the samples to give an approximate 10 nm coating. Samples were then imaged using a Zeiss Supra 55VP located at the Microstructural Analysis Unit at the University of Technology Sydney, using the in-lens detector to collect secondary electrons at 2 kV.
Results and Discussion
The purpose of this study was to investigate the physical event of laser ablation in MALDI. At the time of writing there have been no investigations into how the laser ablation patterns used during IMS impact the sample or even if there is any impact to the data that is acquired. Considering the potential application of IMS to clinical diagnostics, it is important to fully understand what is happening during sample analysis, what this could mean for the resulting data, and if there are any considerations that should be observed when viewing and interpreting the resulting images.
Observations of Laser Ablation
IMS is a non-traditional application of MALDI in the way in which it generates data. Rather than ablating sample in a randomized pattern as the target plate moves, IMS statically ablates individual areas of the sample to generate pixels, moving at predefined increments in a grid pattern. The data can then be viewed by selecting the desired mass and displaying its distribution as a spatial map (Figure 1).
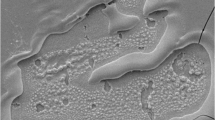
The first consideration is that the target plate does not move as the laser is firing, meaning that more energy is being imparted to the sample at a specific point to generate the ion plume. The assumption that follows is that this results in the complete ablation of the matrix and any co-crystallised analyte at that location leaving no ionizable molecules behind. When it is impractical to adjust the optic of the laser, improving resolution can be achieved through the use of oversampling [7]. When performing this investigation, we chose a 40% oversample, which reduced our ablation spot size from 100 to 60 μm. We have previously published data using this methodology and have found that it is effective at increasing resolution while still maintaining reproducibility [5, 6]. After SEM analysis was completed, we discovered that rather than total ablation, the oversample resulted in a “crescent moon” shaped ablation pattern on the surface of the nitrocellulose (Figure 2).
The most plausible explanation for this is that ionization efficiency of the laser is not uniform; Nd YAG UV lasers conform to a Gaussian energy distribution, which directly translates to a “corona” of light that, at the edge of the beam, does not impart enough energy to initiate an ion plume. In simple terms, only the center ~85% of the laser diameter is capable of completely ablating the sample.
When we measured the diameter of these ablation spots (Figure 3), we found that the edges measured 56 μm across with an internal diameter of approximately 50 μm. This equates to 83% of our expected 60 μm, thereby making our findings consistent with the idea of Gaussian limitations to ablation diameter. We therefore believe that the crescent moon shape is by incomplete UV irradiation of the area resulting in only partial ionization. An intuitive solution to this would be to increase the size of the oversample to ensure that there is a distinct overlap between the areas of total ionization. We acknowledge this and it remains an area of further investigation.
SEM image of an ablation pattern demonstrating an external spot diameter of 56 μm and an internal diameter of 50 μm representing an 83% ablation efficiency of the laser. The single shot laser ablation shape can be seen in Appendix 1
Defining Resolution
Resolution in IMS is traditionally defined as the smallest pixel size achievable by a combination of laser diameter and the size of matrix crystal sizes. In a recent publication, we described the use of sublimation as a matrix application method that creates a homogenous matrix coating, thereby eliminating the gaps that appear during traditional dried droplet or spraying-based preparations [5]. Our investigation of laser ablation and IMS in general has led us to conclude that the actual size of matrix crystals is not important so long as those crystals form a homogenous coverage; it is apparent that the laser possess the ability to partially ablate or “shear” individual crystals and only ionize the part of the sample that is actually being irradiated at the area of highest energy on the Gaussian distribution. Figure 4 shows this, as areas of partial ionization and sheared crystals are clearly visible at the border of the ablation line (indicated by green arrows). With this in mind, we have concluded that the definition of resolution in IMS is somewhat specific to the type of sample preparation used and dependant on a number of key factors. First, resolution should be more correctly described as the diameter of the laser, when applied to a homogenous matrix coating. Small crystal size has been reported to be necessary for high resolution as they create a homogenous coating, with a number of techniques being reported to improve this [8], including addition of matrix dopants [9, 10] and differential rates of drying [11]. However, we have determined this to be a correlational, not causational, relationship; i.e., small crystals can create homogenous coatings but homogenous coating can still be achieved without small crystals. We therefore believe that, so long as no molecules are being delocalized through mechanical disruption or chemical dissolution during sample preparation, crystal size is not a determining factor for resolution. We would also postulate that another contributing factor to resolution is the analyte content within the matrix crystals. Simply put, matrix crystals need to be homogenous and contain enough co-crystallized analyte to generate a sufficient mass signal.
We therefore conclude that the principle factors that need to be considered for increasing resolution are the achievable diameter of the laser and the homogeneity of the matrix coating. This presupposes that sample preparation has been performed in such a way as to prevent delocalization of molecules and maximize the incorporation of matrix and analyte [12]. In the case of droplet-based techniques such as spraying, it is still necessary to define resolution by the average droplet size, as molecules can move freely within the diameter of the droplets; however, for IMS, resolution should be defined as “the homogeneity of the matrix crystals once they have been applied and co-crystallised with the analyte and the effective ablation diameter of the laser.”
Ion Plume “Fallout”
During our SEM analysis of laser ablation, we discovered that there appeared to be a certain amount of unknown debris that is left on the surface of the slide. When we investigated further, we discovered that this debris coated a large proportion of as yet unionized matrix crystals and appeared to be micro crystals of matrix.
At first glance the spots that were already ionized appeared to be relatively clear, not including the crescent moon mentioned earlier (Figure 5).
However, upon closer inspection, these cleared areas appeared to contain debris (Figure 6a) that, when zoomed in, possessed a crystalline structure (Figure 6b).
Following this preliminary finding, we then decided to see how far this debris, which we term “fallout,” was spread through the remainder of the sample slide. When zoomed in on an area of unabated crystal, we found a distinct coating of unknown material (Figure 6a) and that the highest concentration of fallout was at the area directly bordering the ablated area (Figure 7b). We then determined that the concentration of fallout formed a gradient spreading out from the area of ionization (Figure 7c). We therefore concluded that the ion plume was able to throw material a maximum linear distance of 758 μm (Figure 7d).
The discovery of this fallout pattern is, at the time of writing, an undescribed phenomenon that is largely unimportant in traditional MALDI workflows but has several implications for the data collected from IMS. IMS is a technique that is designed to elucidate the physical location of biomolecule within a sample. From our investigation, we have discovered that there is a high likelihood that each laser ablation site contains some material from the previous ablation site; therefore, it is reasonable to assume that molecules are also being spread over an area, thereby “contaminating” the adjacent spot with potentially ionizable molecules. Considering the spatial nature of IMS, this put forth the possibility that ions are being detected that may not actually originate from that area. If this only occurred within a small area, the relatively small amount of material being produced would not be that impactful. However, considering that fallout is “thrown” as far as 700 μm, it is very possible that the spatial maps generated may not actually be true representations of the biology of the sample. Though this fallout is likely very low in concentration, high sensitivity instrumentation such as FTICR or Orbitraps coupled to MALDI sources may indeed detect what is present.
While considering these findings, there is currently no definitive way to measure the contents of fallout nor is there a way of relatively quantifying how the fallout is impacting the spectra being generated. Determining the components of fallout and the concentration of analyte within is currently a future direction and a subject of further investigation.
Conclusion
The purpose of this study was to characterize what is physically happening to a sample that is analyzed via IMS when a grid style laser ablation pattern is used. We discovered that the ablation caused by the laser is not evenly distributed across its diameter and, instead, follows a Gaussian distribution resulting in a corona of incomplete ionization around the outside of the laser beam. It also became apparent that the laser has the ability to “shear” matrix crystals, thereby necessitating a change in the definition of resolution and the factors that control it.
Finally, we also conclude that the “fallout” generated from laser ablation has the potential to introduce error when measuring the spatial distribution of biomolecules. Though this is most likely not an issue with current TOF-based MALDI instruments, as new higher sensitivity instrumentation becomes available and the use of FCTIR and Orbitrap-based mass spectrometry gains popularity, this will likely become an issue that will need resolving.
References
O Rourke, M.B., Djordjevic, S.P.: The renaissance of microbiology: the necessary future for matrix assisted laser desorption ionization mass spectrometry based bio typing. J. Microb. Biochem. Technol. 8, 373–374 (2016)
Karas, M., Hillenkamp, F.: Laser desorption ionization of proteins with molecular masses exceeding 10,000 daltons. Anal. Chem. 60, 2299–2301 (1988)
Caprioli, R.M., Farmer, T.B., Gile, J.: Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal. Chem. 69, 4751–4760 (1997)
Knochenmuss, R.: Ion formation mechanisms in UV-MALDI. Analyst 131, 966–986 (2006)
O'Rourke, M.B., Raymond, B.B.A., Djordjevic, S.P., Padula, M.P.: A versatile cost-effective method for the analysis of fresh frozen tissue sections via matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun. Mass Spectrom. 29, 637–644 (2015)
O'Rourke, M.B., Djordjevic, S.P., Padula, M.P.: A non-instrument-based method for the analysis of formalin-fixed paraffin-embedded human spinal cord via matrix-assisted laser desorption/ionization imaging mass spectrometry. Rapid Commun. Mass Spectrom. 29, 1836–1840 (2015)
Jurchen, J.C., Rubakhin, S.S., Sweedler, J.V.: MALDI-MS imaging of features smaller than the size of the laser beam. J. Am. Soc. Mass Spectrom. 16, 1654–1659 (2005)
O'Rourke, M.B., Djordjevic, S.P., Padula, M.P.: The quest for improved reproducibility in MALDI mass spectrometry. Mass Spectrom. Rev. (2016). doi:10.1002/mas.21515
Kouvonen, P., McDonnell, L.A., Heeren, R.M., Corthals, G.L.: Nitromatrix provides improved LC-MALDI signals and more protein identifications. Proteomics 9, 1662–1671 (2009)
Francese, S., Bradshaw, R., Flinders, B., Mitchell, C., Bleay, S., Cicero, L., Clench, M.R.: Curcumin: a multipurpose matrix for MALDI mass spectrometry imaging applications. Anal. Chem. 85, 5240–5248 (2013)
Cohen, S.L., Chait, B.T.: Influence of matrix solution conditions on the MALDI-MS analysis of peptides and proteins. Anal. Chem. 68, 31–37 (1996)
Bouschen, W., Schulz, O., Eikel, D., Spengler, B.: Matrix vapor deposition/recrystallization and dedicated spray preparation for high-resolution scanning microprobe matrix-assisted laser desorption/ionization imaging mass spectrometry (SMALDI-MS) of tissue and single cells. Rapid Commun. Mass Spectrom. 24, 355–364 (2010)
Author information
Authors and Affiliations
Corresponding author
Appendix 1
Appendix 1

A single shot of the laser showing a circular shape and the ring of partial ionization surrounding the ablated area (indicated by lighter coloured crystals).
Rights and permissions
About this article
Cite this article
O’Rourke, M.B., Raymond, B.B.A. & Padula, M.P. The Characterization of Laser Ablation Patterns and a New Definition of Resolution in Matrix Assisted Laser Desorption Ionization Imaging Mass Spectrometry (MALDI-IMS). J. Am. Soc. Mass Spectrom. 28, 895–900 (2017). https://doi.org/10.1007/s13361-017-1632-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-017-1632-0