Abstract
Ion mobility spectrometry (IMS) coupled with gas-phase hydrogen deuterium exchange (HDX)-mass spectrometry (MS) and molecular dynamic simulations (MDS) has been used for structural investigation of anions produced by electrospraying a sample containing a synthetic peptide having the sequence KKDDDDDIIKIIK. In these experiments the potential of the analytical method for locating charge sites on ions as well as for utilizing collision-induced dissociation (CID) to reveal the degree of deuterium uptake within specific amino acid residues has been assessed. For diffuse (i.e., more elongated) [M – 2H]2– ions, decreased deuterium content along with MDS data suggest that the D4 and D6 residues are charge sites, whereas for the more diffuse [M – 3H]3– ions, the data suggest that the D4, D7, and the C-terminus are deprotonated. Fragmentation of mobility-selected, diffuse [M – 2H]2– ions to determine deuterium uptake at individual amino acid residues reveals a degree of deuterium retention at incorporation sites. Although the diffuse [M – 3H]3– ions may show more HD scrambling, it is not possible to clearly distinguish HD scrambling from the expected deuterium uptake based on a hydrogen accessibility model. The capability of the IMS-HDX-MS/MS approach to provide relevant details about ion structure is discussed. Additionally, the ability to extend the approach for locating protonation sites on positively-charged ions is presented.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The first experiments that combined gas-phase hydrogen deuterium exchange (HDX) and mass spectrometry (MS) as a structural probe for biomolecular ions were conducted more than 20 years ago [1–4]. Shortly thereafter, ion mobility spectrometry (IMS) was combined with HDX-MS to study the reactivities of ions of select mobilities [5, 6]. These studies revealed the maximum HDX levels and rates of exchange for protein and peptide ions and even determined the HDX reactivities of some ions at elevated temperatures [7]. Additionally, it was demonstrated that IMS-HDX-MS data could be used with molecular dynamics simulations (MDS) to provide additional conformational information in the form of hydrogen accessibility [6, 7].
Despite these early demonstrations, IMS-HDX-MS studies were not pursued to a significant degree for nearly 10 years [8]. This may be attributed to several factors. First, the limitations of MDS to obtain reliable structures for large, gas-phase ions hindered the ability to test the various hydrogen accessibility models. Relatedly, the models utilized several speculative assumptions such as charge site locations as well as threshold distances between accessible hydrogens to charge sites to obtain qualitative matches to HDX levels. Therefore, effectively testing such assumptions requires access to accurate structures obtained from MDS. Finally, at the time, site-specific deuterium uptake information to aid model development could not be obtained because of problems associated with hydrogen-deuterium (HD) scrambling during the ion fragmentation process [9, 10].
The nonergodic ion fragmentation techniques of electron capture dissociation (ECD) [11] and electron transfer dissociation (ETD) [12] showed utility early on as a means to locate the sites of deuterium incorporation [13, 14]. Shortly thereafter, HDX-ETD-MS characterization was coupled with ion mobility separations using a traveling wave ion guide (TWIG) instrument [15]. Indeed, using this approach, researchers reported the deuterium uptake for specific ion conformers that were representative of solution conformers for ubiquitin ions. More recently, Rand and coworkers developed a rapid method for gas-phase HDX enabling the analysis of proteins and peptides distinguished by condensed-phase separations [16].
Recently, we reported the combination of a constant-field drift tube with a linear ion trap to perform IMS-HDX-MS/MS experiments [17, 18]. In these studies, dual ion gating allowed the probing of site-specific deuterium uptake for select ion conformations. That is, the gating of mobility-selected ions is conducted prior to their fragmentation (ETD), which is performed in a linear ion trap mass spectrometer. The HDX data was used to develop a model that employed site-specific deuterium uptake data to confirm mobility-matched ion structures obtained from molecular dynamics simulations (MDS) [17]. Later, site-specific HDX kinetics information was used to improve the overall efficiency of the model [18]. Notably, other important studies have used the deuterium uptake kinetics data for mechanistic studies of ion fragmentation [19]. One limitation of these experiments is that linear ion traps employing ETD are largely configured to only allow the fragmentation of positively-charged ions. This complicates studies seeking to determine the accessibility of hydrogens on protein ions for many species. For example, a significant number of proteins contain a disproportionately larger number of acidic residues than basic residues.
One consideration in the study of negatively-charged peptide and protein ions is whether or not collisional activation can be used more effectively (compared with positive ion studies) to ascertain the location of deuterium uptake in IMS-HDX-MS/MS experiments. As part of this consideration it is necessary to briefly review the mechanism of HD scrambling, which can be described by the “mobile proton” model of peptide ion dissociation [20–23]. For positively charged ions, collisional activation leads to intramolecular proton transfer where the locations of protons dictate the types of fragment ions produced. This phenomenon can describe HD scrambling because deuterons can be mobilized as well as protons leading to a shuffling of the deuterium:hydrogen content of specific amino acid residues. The extent of HD scrambling depends not only upon the effective temperature of the ions but also the differences in energy levels of different charge configurations [24]. The same proton mobilization process is required for peptide anion fragmentation [25]. One difference between positive-ion and negative-ion analysis is the source of protons. For the latter ions, these are fewer in number. Additionally, the carboxylic acid side chains result in what has been termed a “locally” mobile proton because of the limited fragment ions formed upon its movement [26]. The consideration of such differences between positively charged and negatively charged ions served as a motivation for the study presented here. Finally, it is noted that some have argued that collision-induced dissociation (CID) should not be used in HDX structural studies because of the potential for HD scrambling [10, 27], while others have suggested some utility for HDX-MS/MS studies [1, 28]. Here we extend the latter consideration by presenting an initial approach that determines the accessibility of hydrogens on model structures obtained from molecular dynamics simulations (MDS).
The experiments described below evaluate the utility of IMS-HDX-MS/MS for the elucidation of structural details for negatively charged peptide ions. Despite the possibility of partial scrambling observed for the peptide ions, site-specific deuterium uptake information can be retained. This uptake information can be used to elucidate structural details such as the location of deprotonation sites as well as the relative accessibilities of hydrogens to those charge sites on the peptide ion structures. Notably, this study extends from previous negative-ion experiments revealing effects of the protein fold as well as charge location on hydrogen site accessibility [29]. This work also lays the foundation for the research described in the accompanying manuscript where limited HD scrambling associated with anions is considered as a potentially enabling phenomenon in the study of protein complex structures.
Experimental
Sample Preparation
The model peptides KKDDDDDIIKIIK and Ace-PAAAAKAAAAKAAAAKAAAAK (Genscript Corp., Piscataway, NJ, USA) were purchased (each >90% purity) and used without further purification. Ultra-pure (chromatography-grade) deionized water, acetonitrile, ammonium hydroxide, and glacial acetic acid (Fisher Scientific, Fair Lawn, NJ, USA) were used to generate stock and ESI solutions of the peptide. The stock solutions consisted of 1.0 mg of peptide in 1.0 mL of ultra-pure water. These solutions were capped in glass vials and subsequently maintained in a refrigerator (4°C). Sample solutions (0.1 mg·mL–1) for ESI were prepared fresh immediately prior to analysis by diluting 0.1 mL of stock solution with the addition of 0.4 mL of ammonium hydroxide (10–2 M) and 0.5 mL of acetonitrile. For the limited positive-ion experiments, for the KKDDDDDIIKIIK peptide, 0.49 mL of stock solution was combined with 0.5 mL of acetonitrile and 0.01 mL of glacial acetic acid. This peptide was selected for these studies because of the potential to study the role of charge inversion (with respect to primary sequence) on the ability to locate deuterium uptake; the generally accepted “relay” mechanism [30, 31] suggests that deuterium incorporation should occur on sites that are accessible to charge sites [6, 7]. For the Ace-PAAAAKAAAAKAAAAKAAAAK solutions, 0.01 mL of glacial acetic acid was added to 0.99 mL of stock solution. ESI solutions were infused (300 nL·min–1) through a pulled-tip capillary (50 μm i.d.) and a bias of –2200 V relative to the instrument entrance orifice was applied to the solution. A bias of +2200 V as used for positive-ion studies. As with the first peptide, this peptide offers the opportunity to study deuterium incorporation as it relates to charge site location.
Ion Mobility Measurements
The history of IMS instrumentation development and IMS applications is well documented in the literature [32–39]. The hybrid instrument combining a constant-field drift tube with a linear ion trap mass spectrometer has also been described previously [17, 18, 29]. A brief description of the instrument, data collection, and analysis are presented here.
Electrosprayed peptide ions were trapped in an ion funnel trap [40, 41] that is housed in a differentially-pumped desolvation chamber and periodically (typically 50 Hz) pulsed into the drift tube. The drift tube was filled with He buffer gas (300 K) and the stacked-ring lenses supported an electric field of ~10 V·cm–1. Ions then separated in the 1-m-long drift tube according to differences in their mobilities through the buffer gas. The second ion gate was activated at specific time delays in order to select ions of specific mobilities for transmission into the linear ion trap (LTQ Velos; ThermoScientific, San Jose, CA, USA) for mass analysis. Delay times between these two gates (front and back of the drift tube) were scanned to generate mobility-resolved MS datasets.
Mass Spectrometry Measurements and the Generation of IMS-MS Datasets
Total ion mass spectra were generated by setting the ion trapping gate and the mobility selection gate to transmit all ions into the mass analyzer. The linear ion trap scan parameters included a m/z range of 80 to 2000 as well as an automatic gain control (AGC) threshold setting of 1 × 106. For these analyses, sample injection times of 200 ms (five microscans) were utilized. During drift time (t D ) selections, the ion gates in the drift tube were activated as mentioned above. Data acquisition was achieved by collecting a single mass spectrum (0.5 min) for each t D selection setting. During ion gating, the AGC was disabled and a sample injection time of 200 ms (five microscans) was also employed. Two-dimensional IMS-MS datasets were generated as described previously [17]. Briefly, an algorithm developed in-house associated each t D selection time with its resultant mass spectrum to create a three-column array file with t D , m/z, and intensity (i) information. From this text file, extracted ion drift time distributions (XIDTDs) [42] and extracted mass spectra could be obtained using user-defined thresholds as described previously [17, 18, 29].
HDX Measurements
Determining the amount of deuterium incorporated into specific peptide ion conformer types using the IMS-MS instrument here has been described in detail previously [17, 18, 29]. Only a brief description is provided here. To accomplish gas-phase HDX, D2O (>99%; Sigma Aldrich, St. Louis, MO, USA) was added to the He buffer gas in the drift tube using a separate leak valve (Granville Philips, Longmont, CO, USA) and monitored using a capacitance manometer (Baratron; MKS, Andover, MA, USA). For these experiments, the He and D2O pressures were typically set at ~2.50 and ~0.04 Torr, respectively; t D distributions were first recorded in pure He to allow the determination of accurate collision cross sections from XIDTDs. The number of deuteriums incorporated into specific ion conformer types were determined by subtracting average m/z values of ions passing through the pure He buffer gas from those obtained after passing through a He:D2O mixture in the drift tube.
The partial pressure of D2O is lower than that used in previous experiments for which maximum HDX levels were reported for peptide anions [29]. In previous studies, kinetics analyses have shown that fast- and slow-exchanging hydrogens are observed for most ions [18]. Previously, it has been suggested that hydrogens located closer to charge sites may comprise the fast-exchanging cohort [6, 7, 29]. To ascertain the relative accessibilities of hydrogens on computer-generated trial structures, a kinetics model is used to represent the fast-exchanging hydrogens (see below). To ensure that the results are not influenced by the more slowly exchanging hydrogens required the use of a lower partial pressure of D2O.
Molecular Dynamics Simulations (MDS)
MDS experiments required the creation of the [M – 3H]3– and [M – 2H]2– KKDDDDDIIKIIK ions. The charge sites for these ions were selected considering ion collision cross sections obtained for MDS structures, gas-phase acidities and Coulomb energies, and deuterium uptake and MS/MS data as described here and in the Results and Discussion section. For [M – 3H]3– peptide ions, the ε-oxygens on residues D4 and D7, and the C-terminus were deprotonated, whereas for [M – 2H]2- ions, the ε-oxygens on residues D4 and D6 were deprotonated. Alternate charge site configurations were also tested for both doubly- and triply-charged KKDDDDDIIKIIK ions; however, these structures typically exhibited collision cross sections that were 20% to 30% larger than experimentally determined values.
Some amino acid residues (neutral K and D residues as well as neutral N- and C-termini) were not predefined in the AMBER force field. For appropriate force field calculations for these residues within the KKDDDDDIIKIIK molecule, two initial, hypothetical structures were utilized. The first structure represented an alpha helix where the φ and ψ bond angles were set to the respective values of –48 and –57. The second structure was an extended conformer where φ and ψ angles of 135 and –140, respectively, were used. Each initial structure was optimized at a quantum chemical level of HF/6–31G(d) and used for calculating molecular electrostatic potential (MEP) values and charge fitting performed with the R.E.D. Server [43–48]. To generate the initial structures for MDS, the AMBER force field FF12SB was employed. The method utilized for simulated annealing was similar to that described previously to generate 1000 candidate structures [49, 50]. The collision cross sections of the 1000 conformers were calculated using the Mobcal [51] software employing the trajectory method (TM) [52]. The potential energies of the 1000 ion conformers obtained from the simulated annealing runs were plotted as a function of collision cross section for each peptide ion. Matching (<2% difference in collision cross section) low-energy ion structures were selected for comparisons to experimental data.
Hydrogen Accessibility Scoring
Deuterium uptake predictions were performed for the KKDDDDDIIKIIK ions using a modified hydrogen accessibility scoring model that has been described in detail previously [17, 18]. In the previous model, the distances between charge sites and heteroatom exchange sites were determined using nominal ion structures obtained from MDS. These exchange sites were then scored based on a summation of the inverse distances to charge sites. Next, the summation of the hydrogen scores for given amino acid residues yielded a residue score. The residue scores were then normalized such that their sum was equal to the experimental deuterium uptake values for the different ion conformers.
For the experiments reported here, these hydrogen accessibility scores served as scaling factors for an “effective” ion-neutral collision model that simulates HDX according to pseudo first order kinetics [18]. As described previously, the ratio of the total number of collisions to the number of effective collisions (resulting in incorporation of deuterium) was estimated based on experimental measurements of ion conformer uptake at low D2O partial pressures. The hydrogen accessibility scores were then used to scale this ratio for individual amino acid residues. Next, using these scaled ratios, a Monte Carlo simulation imposing pseudo first order kinetics for exchange was performed in which the amount of deuterium incorporated for each residue was calculated for a population of ions [18]. Average theoretical uptake values were then compared with the experimentally determined values for each residue to provide an indication of the suitability of the ion structure obtained from MDS.
Results and Discussion
Peptide Ion Collision Cross Sections
Collision cross sections for the dominant features in the t D separation have been determined for all ions of the two samples using the expression [53]:
In Equation 1, ze and k B correspond to the charge of the ion and Boltzmann’s constant, respectively. The variables m I and m B correspond to the mass of the ion and the mass of the buffer gas, respectively. E, L, T, and P correspond to the electric field, length of the drift tube, and the temperature and pressure of the buffer gas, respectively. N represents the neutral number density at standard temperature and pressure (STP) conditions.
Supplementary Figure 1 shows a three-dimensional raised-relief plot of electrosprayed KKDDDDDIIKIIK ions. The most intense features in this dataset are [M – 2H]2– ions. These ions are categorized as two conformer types having t D values of 7.8 ms (Ω = 279 ± 2 Å2) and 9.1 ms (Ω = 315 ± 2 Å2). The more intense (and more diffuse) conformer has an unresolved shoulder at a t D of 8.8 ms. Two dataset features not visible in Supplementary Figure 1 correspond to very low intensity [M – 3H]3– ions. From an XIDTD, the triply-charged ions are shown to exhibit a dominant, diffuse conformer type (Ω = 362 ± 2 Å2) and a less abundant more compact conformer type (Ω = 340 ± 2 Å2) as well as several more diffuse ions of considerably lower abundance. Supplementary Figure 1 also shows the presence of several multimeric species. Because these ions have the same t D values, it is not possible to determine whether or not such species are produced by dissociation of higher-order multimers. In support of this argument is the fact that calculated cross sections for these ions are significantly larger than those determined for many other multimeric peptide ions [54]. Additionally, such multimeric species have been observed previously using this hybrid instrument [29]. Because of this uncertainty, these species are not discussed further.
CID of the Dominant KKDDDDDIIKIIK Ion Conformers and Per-Residue Deuterium Uptake
Supplementary Figure 2 shows the mass spectrum obtained upon CID of the more diffuse [M – 2H]2– KKDDDDDIIKIIK ions. Previous studies have shown that CID of deprotonated peptide ions results in a variety of fragment ions including water loss species as well as internal fragments [55, 56]. Because of this property of anions as well as the loss of ion signal due to mobility selection, the acquisition of the data for the CID mass spectrum requires a significant time. Indeed, the data represented by the mass spectrum shown in Supplementary Figure 2 was collected for 1 h. Among the many different fragment ions produced by CID, a number of ions from which deuterium uptake calculations can be pursued (labeled in Supplementary Figure 2). From this analysis, no ion can be availed to determine the deuterium uptake at the K1 residue. The first ion used is the c2 ion, so here the term “K1 + K2” is used to represent the first two residues of the KKDDDDDIIKIIK peptide.
As mentioned above, collisional activation of precursor ions can cause extensive HD scrambling. One previous study revealed that the use of high-energy CID resulted in near complete HD scrambling for backbone amide sites on deprotonated peptides [10]. It is difficult to directly compare the results presented here to the former studies because instrumental parameters are suggested to affect the degree of HD scrambling. For example, although collision energies must be sufficient to allow for HD scrambling, lower collision energies employed for longer times can lead to significant HD scrambling. Therefore, a note of caution is necessary in that the differences in the results described here can in part result from different instrument parameters.
A comparison of the experimental and theoretical (representing 100% HD scrambling) deuterium content per residue in [M – 2H]2– and [M – 3H]3– KKDDDDDIIKIIK ions is shown in Supplementary Figure 3. Here the per-reside values associated with complete HD scrambling are computed based on the apportioning of the total deuterium content (15.1 and 16 for the doubly and triply charged ions, respectively) according to the fractional number of exchangeable sites on given residues. For the more diffuse [M – 2H]2– ions, a statistically significant difference in deuterium content is observed for residues K1 + K2, D3, D4, D5, D6, and D7. Indeed, only the I8 residue exhibits the same deuterium content value as that expected for complete HD scrambling. Supplementary Figure 3 also shows the comparison that is obtained for [M – 3H]3– KKDDDDDIIKIIK ions. Here residues D3, D4, and to a lesser extent D6 show a disparity when compared with 100% HD scrambling, whereas data for the residues K1 + K2 and D5 are consistent with complete scrambling. That said, the deuterium content values of these residues are also consistent with those obtained from a hydrogen accessibility model indicating the expected gas-phase deuterium uptake for a representative ion conformer type (see discussion below). Having noted the inability to completely eliminate HD scrambling for the [M – 2H]2– and [M – 3H]3– KKDDDDDIIKIIK ions it is necessary to consider the origin of the disparity in deuterium content and that expected for 100% HD scrambling. Notably, the anion CID mechanism [57] and HD scrambling [10] studies for deprotonated peptides suggest that extensive scrambling along the peptide backbone could occur for the KKDDDDDIIKIIK peptide ions. Furthermore, recent IMS-HDX-MS/MS studies show extensive side-chain deuterium uptake relative to backbone deuterium incorporation [17, 18]. The question that arises is whether a property (or properties) of anions could explain the disparity in Supplementary Figure 3. Consider the differences in the numbers and origin of mobile protons for anions and cations. Considerably fewer protons are available for the latter compared with the former. Additionally, the source of protons from carboxylic acid side chains have been described as being “locally” mobile [26, 58]. Taken together, these aspects could account for an overall decrease in proton mobilization leading to a degree of deuterium retention at the incorporation site. That said, such an interpretation should be treated cautiously as instrument parameters have been shown to affect the degree of isotopic scrambling in positively charged ions. As an example, separate experiments have been performed for [M + 3H]3+ ions in which different collisional activation times show differing amounts of HD scrambling via ETD dissociation as shown in Supplementary Figure 4.
Deprotonation Sites on the [M – 2H]2– and [M – 3H]3– KKDDDDDIIKIIK peptide ions
For accurate MDS results, it is necessary to know the location of the deprotonation sites. For the KKDDDDDIIKIIK peptide, there are six possible residues that can represent these sites corresponding to 15 and 20 (for [M – 2H]2– and [M – 3H]3– KKDDDDDIIKIIK ions, respectively) possible arrangements of charges. It may be argued that for MDS simulations, one should consider the apparent gas-phase acidities of different residues [59, 60]; however, estimating apparent gas-phase acidities requires a knowledge of the distance between charge sites [59], which requires candidate structures from MDS. Even knowing the distance between potential charge sites may not be sufficient to accurately estimate correct locations as the most stable ion structure can dictate unique charge arrangements [61].
Here we propose the use of HDX-MS/MS data to find the most likely deprotonation sites for the [M – 2H]2– and [M – 3H]3– KKDDDDDIIKIIK ions. Supplementary Figure 3 shows the per-residue deuterium content after ion fragmentation of the diffuse [M – 2H]2– ions. It can be argued that D residues with a deuterium content greater than 1 should not be assigned as charge sites because they contain more than one exchangeable hydrogen. This leaves three possible charge site locations (residues D4 and D6 and the C-terminus) as well as three different charge site configurations. All three configurations were analyzed by MDS and the charge arrangements, which included the C-terminus yielded structures that were 20% larger (collision cross sections) than the experimental values. Therefore, the D4 and D6 residues were assigned as the deprotonation sites for the [M – 2H]2– KKDDDDDIIKIIK ions. Estimating the deprotonation sites for the [M – 3H]3– KKDDDDDIIKIIK ions using the same approach is straightforward. Only the D4 residue exhibits a deuterium content that is less than 1 (Supplementary Figure 3); therefore, a plausible charge site arrangement consists of residues D4 and D7 and the C-terminus. As with the doubly charged ions, other charge configurations were also modeled and found to lead to structures with generally larger collision cross sections than observed experimentally.
Hydrogen Accessibility Scoring of KKDDDDDIIKIIK Ions and Expected Deuterium Uptake Values
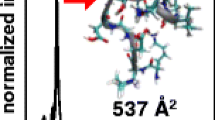
A question arising from the HDX measurements is whether or not the results for the more diffuse [M – 2H]2– and [M – 3H]3– KKDDDDDIIKIIK ions allow the elucidation of structural information. To accomplish this, the deuterium content values can be compared with expected values obtained from a hydrogen scoring model that is applied to nominal ion structures obtained from MDS [17, 18]. Figure 1 shows two ion structures for the diffuse [M – 2H]2– KKDDDDDIIKIIK ions having collision cross sections that match the experimentally determined value. The first ion structure contains deprotonated D4 and D6 residues, whereas the second ion structure contains a deprotonated D3 residue and the C-terminus. A description of the various interactions within the two structures is provided here. For the ion structure with the charge site assignment based on the MS/MS data (Figure 1a), the oxygens on the negatively charged D4 residue are observed to interact with hydrogens on the side chains of the D3 and D5 residues. The deprotonated side chain of the D6 residue is observed to be charge solvated by the I9-K13 peptide backbone. The second ion structure (Figure 1b) exhibits less extensive backbone amide solvation at the C-terminal end. For this structure, the oxygens of the D3 residue interact with the side chain hydrogen of the D5 residue and the backbone amide of the D6 residue. The oxygens on the deprotonated C-terminus interact with the hydrogen on the D6 side chain and the K10 backbone amide. Alternate orientations of these ion structures showing the charge solvation can be found in the Supplementary Information.
Nominal [M – 2H]2– KKDDDDDIIKIIK ion structures obtained from MDS for two different charge site configurations. Both structures exhibit matching collision cross section with the more diffuse (Ω = 315 Å2) [M – 2H]2– KKDDDDDIIKIIK ions. Panel (a) shows a resultant low-energy structure from MDS in which the D4 and D6 residues serve as deprotonation sites, whereas in panel (b) a structure with deprotonated D3 and C-terminus sites is shown. C- and N-termini are labeled as well as select D residues to provide ion structure orientation
The structures obtained from MDS can be used to test the charge site configurations intimated by the MS/MS data. Figure 2 compares the experimentally determined uptake values with theoretical uptake values for individual residues computed using the two [M – 2H]2– ion structures (Figure 1) from MDS and the hydrogen accessibility model. For the ion structure for which the charge site placement suggested by the MS/MS data was utilized, there is generally better agreement with the experiment. For example, the D3, D6, and I8 residues all show a dramatic improvement in the comparison of theory and experiment for this ion structure (Figure 1a). For all other residues, there is no significant difference in predictive capabilities between the two structures.
Bar graphs comparing predicted (red) and experimental (blue) per-residue deuterium uptake values for diffuse [M – 2H]2– KKDDDDDIIKIIK ions. The theoretical uptake values in each panel are calculated using the structures from MDS exhibiting different charge site arrangements (Figure 1). Panel (a) shows the comparison for the ion structure in which the deprotonation sites are selected as the D4 and D6 residues. Panel (b) shows the comparison for the ion structure for which the D3 and C-terminus are used as charge sites. Error bars represent one standard deviation about the mean determined from three measurements
Having noted the superior match with the [M – 2H]2– ion structure shown in Figure 1a, it is instructive to evaluate some of the differences observed between the experimental and theoretical deuterium uptake values (Figure 2a). To consider the effect of complete backbone scrambling, consider the residues with only one exchangeable hydrogen [residues D4 andD6 (charge sites) and I8]. All of these residues have a hydrogen accessibility score that is greater than the average deuterium content. It appears that HD scrambling along the backbone may result in a decrease in deuterium content for all three of these residues. That is, even though these sites are highly accessible to charge sites, HD scrambling results in the movement of incorporated deuteriums to other regions of the peptide.
Having shown the improved agreement (hydrogen accessibility and deuterium content) for the [M – 2H]2– ion structure with the charge locations intimated by the MS/MS data, it is useful to consider the structural information afforded by the per-residue analysis. The D5 residue shows the highest deuterium content value per exchange site (1.7/2) in Figure 2a. Such extensive deuterium content can be ascribed to the location of the D5 residue. That is, the D4 and D6 flanking residues are considered to be charge sites. Therefore, the D5 hydrogens would be more accessible to complex formation involving the charge sites that would be required for exchange via the relay mechanism [30, 31]. Indeed the hydrogen accessibility model also suggests that a higher deuterium content value should be obtained for the D5 residue. Although residues K1 + K2 show a relatively large deuterium content, the ratio of deuterium per exchange site (3/7) is relatively low (Figure 2a). Low deuterium uptake for these residues shows decreased accessibility to the more distal charge sites. Again, this is reflected in the hydrogen accessibility score (3.2). In a separate example, the D3 and D7 residues show about the same deuterium uptake values, which is reflective of their being similarly positioned to single charge sites (the D4 and D6 residues, respectively). The relative deuterium uptake values for the D3 and D7 residues are also accurately captured in the hydrogen accessibility model, even showing a slight increase from the former to the latter residue as determined experimentally (Figure 2a).
Figure 3 shows the two ion structures for the diffuse [M – 3H]3– KKDDDDDIIKIIK ions having matching collision cross sections. The charge site locations for the first and second ion structures are the D4 and D7 residues and the C-terminus and the D3, D5, and D7 residues, respectively. Although these ion structures provide similar collision cross sections, a notable difference is observed. For the first structure with charge placements based on the MS/MS data, the D4 and D7 residues are charge solvated by neighboring D residues as well as the N-terminus. The C-terminus charge protrudes away from the ion. For the second structure, the D5 and D7 residues show a significantly increased degree of charge solvation by the C-terminal residues. Alternate orientations of these ion structures showing the charge solvation can be found in the Supplementary Information.
Nominal [M – 3H]3– KKDDDDDIIKIIK ion structures obtained from MDS for two different charge site configurations. Both structures exhibit matching collision cross section with the more diffuse (Ω = 362 Å2) [M – 3H]3– KKDDDDDIIKIIK ions. Panel (a) shows a resultant low-energy structure from MDS in which the D4 and D7 residues and the C-terminus serve as deprotonation sites, whereas in panel (b) a structure with deprotonated D3, D5, and D7 residues is shown. C- and N-termini are labeled as well as select D residues to provide ion structure orientation
Figure 4 shows the comparison of the hydrogen accessibility scores with the experimental deuterium uptake values for the two [M – 3H]3– KKDDDDDIIKIIK ion structures obtained from MDS. As with the [M – 2H]2– ions, the agreement between experiment and theory is improved for the structure (Figure 3a) for which the charge site location is based upon the MS/MS data (see above). This improvement is observed in the comparison of the D3, D4, and D5 residues. Indeed, only a slight decrease in predictive capability is observed for the K1 + K2 region. Interestingly, the location of the charge site at the D4 residue does not appear to impart an enhancement in the deuterium uptake at neighboring D residues, with the exception perhaps of the D3 residue. For additional comparisons of structures with alternative charge placement see Supplementary Figures 5 and 6. Therefore, although we cannot rule out the possibility that some ions may contain alternate charge sites, the HDX and MDS data are consistent with the charge sites that have been selected.
Bar graphs comparing predicted (red) and experimental (blue) per-residue deuterium uptake values for diffuse [M – 3H]3– KKDDDDDIIKIIK ions. The theoretical uptake values in each panel are calculated using the structures from MDS exhibiting different charge site arrangements (Figure 3). Panel (a) shows the comparison for the ion structure in which the deprotonation sites are selected as the D4 and D7 residues as well as the C-terminus. Panel (b) shows the comparison for the ion structure for which the D3, D5, and D7 residues are used as charge sites. Error bars represent one standard deviation about the mean determined from three measurements
Bar graphs comparing predicted (red) and experimental (blue) per-residue deuterium uptake for the more diffuse [M + 3H]3+ KKDDDDDIIKIIK ions. The charge site arrangement shown in panel (a) is the K1, K10, and K13 residues. For panel (b) the charge site arrangement is the K1, K2, and K13 residues. Error bars represent one standard deviation about the mean determined from three measurements. The experimental data are from reference [17]
Bar graphs comparing predicted (red) and experimental (blue) per-residue deuterium uptake for the more diffuse [M + 3H]3+ Ace-PAAAAKAAAAKAAAAKAAAAK ions. The charge site arrangement shown in panel (a) is the K6, K16, and K21 residues. For panel (b) the charge site arrangement is the K6, K11, and K21 residues. Error bars represent one standard deviation about the mean determined from three measurements
Extension of Charge Site Localization to Positively-Charged Ions
Although this proof-of-concept study shows an advantage for IMS-HDX-MS/MS in determining the locations of charges for negatively charged peptide ions, it is desirable to test the applicability to other types of ions. Here we compare results for positively charged peptide ions. Prior IMS-HDX-MS/MS (via ETD) studies of [M + 3H]3+ KKDDDDDIIKIIK ions have provided candidate structures from MDS, which show good agreement between experimental and theoretical deuterium uptake values [17]. For the matching ion structure, the K1, K10, and K13 residues are protonated. Figure 5a shows that the good agreement between the hydrogen accessibility method [17] and the experimental deuterium uptake. Indeed, the theory captures the relative changes in deuterium uptake between residues remarkably well with the exception of the K10 and I12 residues. For a separate ion structure of matching collision cross section having protonation sites of K1, K2, and K10, the agreement is markedly decreased (Figure 5b). Indeed the only residues that do show agreement between the experiment and the theory are the D7, I8, and I11 residues.
To further demonstrate the utility of the IMS-HDX-MS/MS in assigning charge sites in positive ion mode, separate experiments were performed for peptide ions formed by electrospraying the Ace-PAAAAKAAAAKAAAAKAAAAK peptide. As with the positively charged KKDDDDDIIKIIK ions, the MS/MS analysis is performed by ETD in order to preserve the location of the deuterium label. After HDX, the more compact (Ω = 417 Å2) [M + 3H]3+ Ace-PAAAAKAAAAKAAAAKAAAAK ions were mobility selected and subjected to ETD. The per-residue deuterium content for the compact conformer is shown in Figure 6.
Selecting three charge locations from four available sites results in four possible charge site configurations. The observation of the c6 and z1 ions in the ETD spectra suggests that the K6 and K21 residues are protonation sites. The question that arises is whether the K11 or the K16 residue is the remaining protonation site. The potential charge arrangements (residues K6, K11, K21, and K6, K16, K21) were used in MDS. Figure 6a shows the predicted deuterium uptake pattern for the mobility-matched structure using residues K6, K11, and K21 as protonation sites. A comparison with experimental deuterium uptake values shows good agreement with data for this charge site assignment with the exception of residues A17, A18, and A20. For residues K6, K11, and K16, a smaller deviation is observed (<0.3). Figure 6b compares the predicted deuterium uptake pattern for the mobility-matched structure with the K6, K16, and K21 residue charge site arrangement with the experimental values. Residues K11, K16, and K21 exhibit a significant deviation (>0.7) from the experimental deuterium uptake values. Therefore, these data suggest that the appropriate charge site assignment is residues K6, K11, and K16.
Conclusions
The deuterium uptake levels have been recorded for select ion conformations of negatively charged peptide ions. CID of [M – 2H]2– and [M – 3H]3– KKDDDDDIIKIIK peptide ions suggests only a limited degree of HD scrambling; label retention is observed for some amino acid residues that appear to be sites of incorporation. Although the utility of solution HDX with MS/MS for peptide anions has been called into question [10], the work here presents a novel experimental utility. One example is the ability to use the partial retention information with the IMS measurements to support candidate ion conformations obtained from MDS. Another advantage of the partial label retention demonstrated here is the ability to locate charge sites based on comparisons of per-residue deuterium incorporation. These charge site determinations aid the production of candidate ion structures from MDS, which are necessary to obtain useful structural information from IMS-HDX data in the form of hydrogen site accessibility. Although several drawbacks exist for these negative ion analyses including the potential for HD scrambling and the production of more complicated ion fragmentation spectra, these proof-of-concept experiments show that the mapping of charge sites and exchange sites with CID is possible to some degree for negatively charged ions. Additionally, the approach can be extended to positively charged peptide ions to confirm the locations of assigned protonation sites for MDS.
References
Smith, D.L., Zhang, Z.Q., Liu, Y.Q.: Amide hydrogen-exchange and mass-spectrometry - a probe of high-order structure in proteins. Pure Appl. Chem. 66, 89–94 (1994)
Katta, V., Chait, B.T.: Conformational-changes in proteins probed by hydrogen-exchange electrospray-ionization mass-spectrometry. Rapid Commun. Mass Spectrom. 5, 214–217 (1991)
Katta, V., Chait, B.T.: Hydrogen-deuterium exchange electrospray-ionization mass-spectrometry - a method for probing protein conformational-changes in solution. J. Am. Chem. Soc. 115, 6317–6321 (1993)
Suckau, D., Shi, Y., Beu, S.C., Senko, M.W., Quinn, J.P., Wampler, F.M., McLafferty, F.W.: Coexisting stable conformations of gaseous protein ions. Proc. Natl. Acad. Sci. U. S. A. 90, 790–793 (1993)
Valentine, S.J., Clemmer, D.E.: H/D exchange levels of shape-resolved cytochrome c conformers in the gas phase. J. Am. Chem. Soc. 119, 3558–3566 (1997)
Wyttenbach, T., Bowers, M.T.: Gas phase conformations of biological molecules: The hydrogen/deuterium exchange mechanism. J. Am. Soc. Mass Spectrom. 10, 9–14 (1999)
Valentine, S.J., Clemmer, D.E.: Temperature-dependent H/D exchange of compact and elongated cytochrome c ions in the gas phase. J. Am. Soc. Mass Spectrom. 13, 506–517 (2002)
Bohrer, B.C., Atlasevich, N., Clemmer, D.E.: Transitions between elongated conformations of ubiquitin [M + 11H]11+ enhance hydrogen/deuterium exchange. J. Phys. Chem. B 115, 4509–4515 (2011)
Deng, Y.Z., Pan, H., Smith, D.L.: Selective isotope labeling demonstrates that hydrogen exchange at individual peptide amide linkages can be determined by collision-induced dissociation mass spectrometry. J. Am. Chem. Soc. 121, 1966–1967 (1999)
Bache, N., Rand, K.D., Roepstorff, P., Ploug, M., Jorgensen, T.J.D.: Hydrogen atom scrambling in selectively labeled anionic peptides upon collisional activation by MALDI tandem time-of-flight mass spectrometry. J. Am. Soc. Mass Spectrom. 19, 1719–1725 (2008)
Zubarev, R.A., Kelleher, N.L., McLafferty, F.W.: Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc. 120, 3265–3266 (1998)
Syka, J.E.P., Coon, J.J., Schroeder, M.J., Shabanowitz, J., Hunt, D.F.: Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U. S. A. 101, 9528–9533 (2004)
Rand, K.D., Adams, C.M., Zubarev, R.A., Jorgensen, T.J.D.: Electron capture dissociation proceeds with a low degree of intramolecular migration of peptide amide hydrogens. J. Am. Chem. Soc. 130, 1341–1349 (2008)
Rand, K.D., Pringle, S.D., Morris, M., Brown, J.M.: Site-specific analysis of gas-phase hydrogen/deuterium exchange of peptides and proteins by electron transfer dissociation. Anal. Chem. 84, 1931–1940 (2012)
Rand, K.D., Pringle, S.D., Murphy, J.P., Fadgen, K.E., Brown, J., Engen, J.R.: Gas-phase hydrogen/deuterium exchange in a traveling wave ion guide for the examination of protein conformations. Anal. Chem. 81, 10019–10028 (2009)
Mistarz, U.H., Brown, J.M., Haselmann, K.F., Rand, K.D.: Simple setup for gas-phase H/D exchange mass spectrometry coupled to electron transfer dissociation and ion mobility for analysis of polypeptide structure on a liquid chromatographic time scale. Anal. Chem. 86, 11868–11876 (2014)
Khakinejad, M., Kondalaji, S.G., Maleki, H., Arndt, J.R., Donohoe, G.C., Valentine, S.J.: Combining ion mobility spectrometry with hydrogen-deuterium exchange and top-down MS for peptide ion structure analysis. J. Am. Soc. Mass Spectrom. 25, 2103–2115 (2014)
Khakinejad, M., Kondalaji, S., Tafreshian, A., Valentine, S.: Gas-phase hydrogen-deuterium exchange labeling of select peptide ion conformer types: a per-residue kinetics analysis. J. Am. Soc. Mass Spectrom. 26, 1115–1127 (2015)
Fattahi, A., Zekavat, B., Solouki, T.: H/D Exchange kinetics: experimental evidence for formation of different b fragment ion conformers/isomers during the gas-phase peptide sequencing. J. Am. Soc. Mass Spectrom. 21, 358–369 (2010)
Jones, J.L., Dongre, A.R., Somogyi, A., Wysocki, V.H.: Sequence dependence of peptide fragmentation efficiency curves determined by electrospray-ionization surface-induced dissociation mass-spectrometry. J. Am. Chem. Soc. 116, 8368–8369 (1994)
Dongre, A.R., Jones, J.L., Somogyi, A., Wysocki, V.H.: Influence of peptide composition, gas-phase basicity, and chemical modification on fragmentation efficiency: evidence for the mobile proton model. J. Am. Chem. Soc. 118, 8365–8374 (1996)
Burlet, O., Orkiszewski, R.S., Ballard, K.D., Gaskell, S.J.: Charge promotion of low-energy fragmentations of peptide ions. Rapid Commun. Mass Spectrom. 6, 658–662 (1992)
Harrison, A.G., Yalcin, T.: Proton mobility in protonated amino acids and peptides. Int. J. Mass Spectrom. 165, 339–347 (1997)
Zhang, Z.Q.: Prediction of low-energy collision-induced dissociation spectra of peptides with three or more charges. Anal. Chem. 77, 6364–6373 (2005)
Grzetic, J., Oomens, J.: Effect of the Asn side chain on the dissociation of deprotonated peptides elucidated by IRMPD spectroscopy. Int. J. Mass Spectrom. 354, 70–77 (2013)
Wysocki, V.H., Tsaprailis, G., Smith, L.L., Breci, L.A.: Special feature: commentary— mobile and localized protons: a framework for understanding peptide dissociation. J. Mass Spectrom. 35, 1399–1406 (2000)
Hamuro, Y., Tomasso, J.C., Coales, S.J.: A simple test to detect hydrogen/deuterium scrambling during gas-phase peptide fragmentation. Anal. Chem. 80, 6785–6790 (2008)
Xiao, H., Kaltashov, I.A.: Transient structural disorder as a facilitator of protein-ligand binding: native H/D exchange-mass spectrometry study of cellular retinoic acid binding protein I. J. Am. Soc. Mass Spectrom. 16, 869–879 (2005)
Donohoe, G.C., Khakinejad, M., Valentine, S.J.: Ion mobility spectrometry-hydrogen deuterium exchange mass spectrometry of anions: Part 1. Peptides to proteins. J. Am. Soc. Mass Spectrom. 26, 564–576 (2015)
Campbell, S., Rodgers, M.T., Marzluff, E.M., Beauchamp, J.L.: Deuterium exchange reactions as a probe of biomolecule structure. Fundamental studies of cas phase H/D exchange reactions of protonated glycine oligomers with D2O, CD3OD, CD3CO2D, and ND3. J. Am. Chem. Soc. 117, 12840–12854 (1995)
Chan, S., Enke, C.G.: Mechanistic study of hydrogen-deuterium exchange between M-1- ions of chlorinated benzenes and D2O OR ND3. J. Am. Soc. Mass Spectrom. 5, 282–291 (1994)
Stlouis, R.H., Hill, H.H.: Ion mobility spectrometry in analytical-chemistry. Crit. Rev. Anal. Chem. 21, 321–355 (1990)
Clemmer, D.E., Jarrold, M.F.: Ion mobility measurements and their applications to clusters and biomolecules. J. Mass Spectrom. 32, 577–592 (1997)
Woods, L.A., Radford, S.E., Ashcroft, A.E.: Advances in ion mobility spectrometry-mass spectrometry reveal key insights into amyloid assembly. Biochim. Biophys. Acta. 1834, 1257–1268 (2013)
Wyttenbach, T., Pierson, N.A., Clemmer, D.E., Bowers, M.T.: Ion mobility analysis of molecular dynamics. Annu. Rev. Phys. Chem. 65, 175–196 (2014)
Maurer, M.M., Donohoe, G.C., Valentine, S.J.: Advances in ion mobility-mass spectrometry instrumentation and techniques for characterizing structural heterogeneity. Analyst 140, 6782–6798 (2015)
Hyung, S.-J., Ruotolo, B.T.: Integrating mass spectrometry of intact protein complexes into structural proteomics. Proteomics 12, 1547–1564 (2012)
May, J.C., McLean, J.A.: Ion mobility-mass spectrometry: time-dispersive instrumentation. Anal. Chem. 87, 1422–1436 (2015)
Cumeras, R., Figueras, E., Davis, C.E., Baumbach, J.I., Gracia, I.: Review on ion mobility spectrometry. Part 1: current instrumentation. Analyst 140, 1376–1390 (2015)
Donohoe, G.C., Maleki, H., Arndt, J.R., Khakinejad, M., Yi, J.H., McBride, C., Nurkiewicz, T.R., Valentine, S.J.: A new ion mobility-linear ion trap instrument for complex mixture analysis. Anal. Chem. 86, 8121–8128 (2014)
Tang, K., Shvartsburg, A.A., Lee, H.N., Prior, D.C., Buschbach, M.A., Li, F.M., Tolmachev, A.V., Anderson, G.A., Smith, R.D.: High-sensitivity ion mobility spectrometry/mass spectrometry using electrodynamic ion funnel interfaces. Anal. Chem. 77, 3330–3339 (2005)
Lee, S., Li, Z.Y., Valentine, S.J., Zucker, S.M., Webber, N., Reilly, J.P., Clemmer, D.E.: Extracted fragment ion mobility distributions: a new method for complex mixture analysis. Int. J. Mass Spectrom. 309, 154–160 (2012)
Vanquelef, E., Simon, S., Marquant, G., Garcia, E., Klimerak, G., Delepine, J.C., Cieplak, P., Dupradeau, F.-Y.: RED Server: a web service for deriving RESP and ESP charges and building force field libraries for new molecules and molecular fragments. Nucleic Acids Res. 39, W511–W517 (2011)
The PyRED (or R.E.D. Python) program:F. Wang, J.-P. Becker, P. Cieplak & F.-Y. Dupradeau, R.E.D. Python: Object oriented programming for Amber force fields, Université de Picardie - Jules Verne, Sanford Burnham Prebys Medical Discovery Institute, Nov. 2013
Dupradeau, F.Y., Pigache, A., Zaffran, T., Savineau, C., Lelong, R., Grivel, N., Lelong, D., Rosanski, W., Cieplak, P.: The R.ED. tools: advances in RESP and ESP charge derivation and force field library building. Phys. Chem. Chem. Phys. 12, 7821–7839 (2010)
Bayly, C.I., Cieplak, P., Cornell, W.D., Kollman, P.A.: A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges—the RESP model. J. Phys. Chem. 97, 10269–10280 (1993)
Schmidt, M.W., Baldridge, K.K., Boatz, J.A., Elbert, S.T., Gordon, M.S., Jensen, J.H., Koseki, S., Matsunaga, N., Nguyen, K.A., Su, S.J., Windus, T.L., Dupuis, M., Montgomery, J.A.: General atomic and molecular electronic-structure system. J. Comput. Chem. 14, 1347–1363 (1993)
Gordon, M.S., Schmidt, M.W.: In: Dykstra, C.E., Frenking, G., Kim, K.S., Scuseria, G.E. (eds.) Advances in electronic structure theory: GAMESS a decade later. Elsevier, Amsterdam (2005)
Gidden, J., Bowers, M.T.: Gas-phase conformational and energetic properties of deprotonated dinucleotides. Eur. Phys. J. D 20, 409–419 (2002)
Wyttenbach, T., von Helden, G., Bowers, M.T.: Gas-phase conformation of biological molecules: Bradykinin. J. Am. Chem. Soc. 118, 8355–8364 (1996)
Jarrold Research Group, I.U. MOBCAL – A Program to Calculate Mobilities. [Cited 2015 August 17]; Available from: http://www.indiana.edu/~nano/software.html
Mesleh, M.F., Hunter, J.M., Shvartsburg, A.A., Schatz, G.C., Jarrold, M.F.: Structural information from ion mobility measurements: effects of the long-range potential. J. Phys. Chem. 100, 16082–16086 (1996)
Mason, E.A., Mcdaniel, E.W.: Transport properties of ions in gases. John Wiley & sons, New York (1988)
Counterman, A.E., Valentine, S.J., Srebalus, C.A., Henderson, S.C., Hoaglund, C.S., Clemmer, D.E.: High-order structure and dissociation of gaseous peptide aggregates that are hidden in mass spectra. J. Am. Soc. Mass Spectrom. 9, 743–759 (1998)
Brinkworth, C.S., Dua, S., McAnoy, A.M., Bowie, J.H.: Negative ion fragmentations of deprotonated peptides: backbone cleavages directed through both Asp and Glu. Rapid Commun. Mass Spectrom. 15, 1965–1973 (2001)
Steinborner, S.T., Bowie, J.H.: A comparison of the positive- and negative-ion mass spectra of bio-active peptides from the dorsal secretion of the Australian red tree frog. Litoria rubella. Rapid Commun. Mass Spectrom. 10, 1243–1247 (1996)
Harrison, A.G.: Sequence-specific fragmentation of deprotonated peptides containing H or alkyl side chains. J. Am. Soc. Mass Spectrom. 12, 1–13 (2001)
Gu, C.G., Tsaprailis, G., Breci, L., Wysocki, V.H.: Selective gas-phase cleavage at the peptide bond terminal to aspartic acid in fixed-charge derivatives of asp-containing peptides. Anal. Chem. 72, 5804–5813 (2000)
Schnier, P.D., Gross, D.S., Williams, E.R.: On the maximum charge-state and proton-transfer reactivity of peptide and protein ions formed by electrospray-ionization. J. Am. Soc. Mass Spectrom. 6, 1086–1097 (1995)
Zhang, X., Cassady, C.J.: Apparent gas-phase acidities of multiply protonated peptide ions: Ubiquitin, insulin B, and renin substrate. J. Am. Soc. Mass Spectrom. 7, 1211–1218 (1996)
Kohtani, M., Jones, T.C., Sudha, R., Jarrold, M.F.: Proton transfer-induced conformational changes and melting in designed peptides in the gas phase. J. Am. Chem. Soc. 128, 7193–7197 (2006)
Acknowledgments
The authors gratefully acknowledge financial support from the Eberly College of Arts and Sciences at West Virginia University for providing laboratory research startup funds. Additionally, partial support for this work was provided by the National Institutes of Health (1R15NS090380-01 and 1R01GM114494-01).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 2734 kb)
Rights and permissions
About this article
Cite this article
Khakinejad, M., Ghassabi Kondalaji, S., Donohoe, G.C. et al. Ion Mobility Spectrometry-Hydrogen Deuterium Exchange Mass Spectrometry of Anions: Part 2. Assessing Charge Site Location and Isotope Scrambling. J. Am. Soc. Mass Spectrom. 27, 451–461 (2016). https://doi.org/10.1007/s13361-015-1304-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-015-1304-x