Abstract
We describe a novel technique combining precise organelle microextraction with deposition and matrix-assisted laser desorption/ionization (MALDI) for a rapid, minimally invasive mass spectrometry (MS) analysis of single organelles from living cells. A dual-positioner nanomanipulator workstation was utilized for both extraction of organelle content and precise co-deposition of analyte and matrix solution for MALDI-direct organelle mass spectrometry (DOMS) analysis. Here, the triacylglycerol (TAG) profiles of single lipid droplets from 3T3-L1 adipocytes were acquired and results validated with nanoelectrospray ionization (NSI) MS. The results demonstrate the utility of the MALDI-DOMS technique as it enabled longer mass analysis time, higher ionization efficiency, MS imaging of the co-deposited spot, and subsequent MS/MS capabilities of localized lipid content in comparison to NSI-DOMS. This method provides selective organellar resolution, which complements current biochemical analyses and prompts for subsequent subcellular studies to be performed where limited samples and analyte volume are of concern.

ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The development of analytical techniques and instrumentation for single cell organelle analysis by mass spectrometry has been the focus of many groups over the last decade. Chemical differences of each organelle have been well documented in several biological systems because of the known heterogeneity of individual cells and organelles [1–5]. Several separation technologies have been implemented to enable single cell studies, such as microfluidic devices, capillary electrophoresis, flow cytometry [4, 6], laser capture microdissection [7, 8], and printed microarrays [9] to combat the inherent population averaging obtained from typical multiple cell analyses. However, these methods suffer from extensive extraction protocols and sample preparation. They also commonly require expenditure of an entire sample of cells, preventing the possibility for additional analysis of the same tissue or culture sample.
We have developed a nanomanipulation workstation capable of collecting trace particles [10–13], single cells, and subcellular organelles [5, 14, 15]. The workstation can be equipped with up to four piezoelectric nanopositioners capable of simultaneously utilizing different end effectors, including microgrippers, nanospray emitters, and microelectrodes that perform low impedance electrical characterizations. The joystick-controlled nanopositioners have 12 mm range of motion in the X and Z axes, and 28 mm range in the Y axis. The translational resolution of the nanomanipulator allows for 100 and 5 nm in coarse and fine mode, respectively. Owing to the precise targeting capability, extractions are performed discriminately, removing the need for lengthy chromatographic separations and simultaneously reducing matrix effects [12, 13]. This allows extractions to be introduced directly into the mass spectrometer via direct infusion mass spectrometry (DIMS), which reduces additional sample preparation and enables rapid mass analysis. Using DIMS in combination with NSI-MS has several advantages, including small analyte concentration requirement (pmol/μL), the removal of prerequisite sheath gas, and improved ionization efficiency [16].
Recently, we have shown that the nanomanipulator can be used as a surface-probing technique, direct analyte-probed nanoextraction (DAPNe), to extract drug residue from latent fingerprints [10], individual fibers [13], and trace explosive materials [12], in combination with direct analysis in real time mass spectrometry (DART-MS) [11]. DAPNe has also been utilized for the analysis of document inks, demonstrating the ability to characterize the chemistry of both antique and modern documents without interfering with the integrity of the document [17]. The translational resolution of the nanomanipulator has been particularly useful for the biological investigation and analysis of single cells and organelles. In isolated and purified lipid droplets (i.e., released from several cells) from wild type and transgenic cottonseed embryos, NSI-DOMS revealed unexpected organelle-to-organelle heterogeneity of triacylglycerol (TAG) molecular species [5]. This discovery led to the single cell and organelle investigation of other cell types with the nanomanipulator. In human breast tissue, cancer associated adipocytes (CAAs) and adjacent healthy adipocytes revealed distinct differences in heterogeneity of TAG molecular species composition from extractions using a two-tip method with the nanomanipulator using NSI [14]. Vastly different lipid profiles were also obtained dependent on lipid droplet size from whole cell digestions of differentiated human adipocytes using NSI, which validated the workstation as a valuable tool for in vitro cell cultures [18]. Using microextractions and NSI for biological work has proven to be advantageous in several experiments, but is not without issues and areas for improvement. Positioning of the nanospray capillary can greatly affect the resulting spectra obtained, especially in cases where the extracted analyte is concentrated near the end of the tip. The fragility of the nanospray emitter opening can be affected and damaged during the process of extraction by coming into contact with culture dishes and slides, which influences the resulting spray during ionization. Most importantly, the small volume of extracted cell content greatly limits analysis time with the sample. It is for these reasons that a different source for ionizing and analyzing extracted cellular content was desired, leading to the work presented here. MALDI mass spectrometry imaging (MALDI-MSI) has become revolutionary in mapping lipids [19], proteins [20], and other small molecules [21] in tissues, and additionally in combination with liquid chromatography [22]. However, its spatial resolution for single cell and organelle imaging has been limited by dispersement of analytes during spotting of matrix and laser spot size [23]. Here, we introduce a novel technique to provide an alternative solution to the problem, by coupling the microextraction capabilities of the nanomanipulation workstation with a co-deposition method for use in MALDI MS analysis using a high-resolution, accurate mass (<3 ppm [24]) hybrid MALDI-LTQ-XL-Orbitrap mass spectrometer. The nanomanipulator was equipped with two nanopositioners, each fitted with a nanospray emitter; one for the direct microextraction and deposition of individual lipid droplets and the second for matrix application (Figure 1). As a proof-of-concept experiment, we extracted individual, single lipid droplets from adipocytes that were differentiated from 3T3-L1 murine fibroblasts. The lipid-droplet microextracts were collected on-stage, spotted onto glass slides, and coated with a droplet of 2,5-dihydroxybenzoic acid (DHB) matrix solution, all achieved using the nanomanipulation platform. Prepared samples were immediately analyzed using a MALDI-LTQ-XL-Orbitrap at 100,000 resolution (at m/z 400). Individual lipid droplet triacylglycerol (TAG) profiles were acquired from full MS and MS/MS scans and were compared with the relative distribution of TAG lipid profiles collected using nanoelectrospray ionization mass spectrometry (NSI-MS) on a linear ion trap LTQ XL mass spectrometer. There have been similar attempts at single cell [25–31] and organelle [32–34] analyses from tissues and cell cultures using both ESI/NSI and MALDI that merit significant recognition. However, each technique has its own advantages as well as a variety of shortcomings, such as nondiscriminate extractions [26, 30, 31], limited time with samples and poor sensitivity [32], additional purification or digestion steps [25, 26, 31, 33], and expenditure of multiple cells [7, 26, 33, 34]. Rubakhin et al. [33] manipulated individual organelles onto a glass coverslip for MALDI analysis, but this method required liberation of organelles from multiple cells, losing spatial information, an additional washing step, and consumed the entire sample during a single laser ablation event, preventing MS/MS analysis.
The technique described here provides a rapid, discriminate in vitro microextraction and analytical method for the analysis of single organelles without the consumption of multiple cells or additional purification and digestion steps. Using nanomanipulation-coupled MALDI-DOMS, we were able to achieve a minimum of 15-fold increase in analysis time per sample, up to three orders of magnitude higher ionization efficiency, and subsequent MS/MS capabilities following MSI of the co-deposited spot when compared to NSI-DOMS.
Experimental
Adipogenic Differentiation
Murine fibroblasts 3T3-L1 (ATCC, Manassas, VA, USA) were seeded in 33 mm glass bottom culture dishes (Matek, Ashland, MA, USA) at 15,000 cells per dish. Growth medium consisted of 3 mL Dulbecco’s modified Eagle’s medium (DMEM) containing 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (all from Sigma-Aldrich, St. Louis, MO, USA). Adipogenic differentiation was carried out using previously described methods [35–37]. Upon reaching 100% confluence (5 d), growth medium was removed and replaced with 3 mL differentiation medium consisting of DMEM with 10% FBS, 1% penicillin-streptomycin, 0.5 mM 3-isobutyl-1-methylxanthine (IBMX), 1 μM dexamethasone, 10 μg/mL insulin, 0.2 mM indomethacin, and either 100 μM of palmitic acid or oleic acid to promote lipid droplet growth [36] (all from Sigma-Aldrich, St. Louis, MO, USA). Differentiation medium was replaced every 2 d until lipid droplets were observed, approximately 14 d. When lipid droplets reached between 20 and 50 μm, differentiation medium was removed from the culture dish and replaced with 2 mL of 1× phosphate buffered saline (PBS) (Fisher Scientific, Fair Lawn, NJ, USA) in preparation for microextraction. The culture plates containing the differentiated adipocytes were then transferred to the nanomanipulator for microextraction.
Microextraction of Lipid Droplets and Matrix Deposition
An L200 nanomanipulator workstation (DCG Systems Inc., Fremont, CA, USA) was mounted on a TE-2000 inverted microscope (Nikon, Melville, NJ, USA) and equipped with a four-channel, 60 psi pressure injector connected to a nitrogen gas cylinder (MicroData Instrument Inc., S. Plainfield, NJ, USA). The nanomanipulator was equipped with two nanopositioners, each holding a 1.0 ± 0.2 μm diameter palladium-gold coated nanospray emitter (New Objective, Woburn, MA, USA). Emitters for lipid droplet extractions were back-loaded with 10 μL of 2:1 chloroform:methanol (v/v). Emitter solvents for NSI-MS experiments were prepared with 0.1% ammonium acetate. Emitters for matrix deposition were back loaded with 10 μL of a 20 mg/mL DHB solution in 3:2 acetonitrile:water (v/v) (DHB 98%; and all solvents HPLC grade from Sigma Aldrich, St. Louis, MO, USA). The co-deposition method using the nanomanipulation platform was tested in 6 replicates by backfilling and spotting a nanospray emitter with 250 μg/mL TAG standard (17:0/17:0/17:0) (Sigma Aldrich, St. Louis, MO, USA) in chloroform, followed by matrix deposition with the second nanopositioner (Figure 2).
Using the control joystick, the nanospray extraction emitter was directed towards a selected lipid droplet and positioned within the lipid droplet (Figure 3a). The contents of the respective lipid droplet were aspirated into the tip using 20 psi from the pressure injector (Online Resource 1). The emitter was then transitioned out of the culture dish and the culture dish removed. The nanospray emitter containing the lipid droplet was either: (a) removed and directly transferred to the NSI source for NSI-MS analysis or (b) was brought into contact with a glass slide and was ejected from the emitter at a pressure of 1.5 psi for approximately 0.3 s. The solvent was allowed to evaporate and was pulse-spotted approximately 20 additional times (Figure 3b). The second emitter containing matrix solution was then positioned on top of the analyte spot and ejected from the emitter at a pressure of 1.5 psi, until the original spot was fully covered with matrix (Figure 3c and Online Resource 2). Experiments were performed in four replicates (n = 4) for both NSI and MALDI experiments.
(a) Nanospray emitter positioned within lipid droplet of a 3T3-L1 differentiated adipocyte in preparation for microextraction. (b) Following microextraction, the nanospray emitter (top) is shown depositing the dissolved lipid droplet contents onto a precleaned glass slide, the second emitter (left) is utilized to spot matrix solution on top of the analyte spot, resulting in an ~450 μm matrix spot (c). Scale bars represent 50 μm
MALDI-LTQ-Orbitrap Analysis
MALDI-DOMS experiments were conducted on a MALDI-LTQ-XL-Orbitrap (Thermo Scientific, San Jose, CA, USA). The instrument was equipped with an intermediate pressure MALDI source (75 mTorr) using a 337 nm N2 laser (MNL 100; Lasertechnik Berlin, Germany). Instrument experimental conditions were as follows: 12 μJ energy per laser shot, 10 laser shots per spectra, laser raster motion was set at a 50 μm step size. Data acquisition was collected in positive ion mode using a mass range of m/z 700–1000 and mass resolution of 100,000 (at m/z 400). MS/MS experimental CID conditions were: 1.0 u isolation width, 30.0% collision energy (arb), and 40.0 ms activation time.
NSI LTQ Analysis
NSI-MS experiments were conducted on an LTQ XL linear ion trap mass spectrometer (Thermo Scientific, San Jose, CA, USA) equipped with a NSI source (Proxeon Biosystems, Odense, Denmark). Instrument experimental conditions were as follows: spray voltage of 2.5 kV, with 200°C capillary temperature and capillary voltage of 9 V. Scans were analyzed in positive mode with a mass range of m/z 100–1000.
Data Analysis
Spectra processing for both NSI and MALDI experiments was performed using Xcalibur v 2.2 (Thermo Scientific, San Jose, CA, USA). Image processing for the co-deposited spots was performed with ImageQuest v 1.01 (Thermo Scientific, San Jose, CA, USA). Spectra were plotted using PSI-Plot (Poly Software International, Pearl River, NY, USA).
Results and Discussion
Nanomanipulator Single Organelle Extraction and Deposition
The precise translational resolution of the nanopositioners allowed the exact positioning of the emitter tip within single lipid droplets, ensuring that during the extraction only the lipid droplet and its contents were aspirated (Figure 3a and Online Resource 1). In the emitter, the lipid droplet becomes dispersed in extraction solvent and is primarily concentrated at the emitter tip. This proved to be beneficial during analyte spotting for MALDI-DOMS, as the dissolved sample was expelled from the emitter within the first microliter spotted. Spotting the lipid droplet was achieved using a pulse-spotting method, where small sub-microliter amounts of dissolved analyte were spotted onto the slide, solvent was allowed to evaporate, and the process was repeated approximately 20 times. The combination of the pulse-spotting method and analyte concentration in the emitter tip yielded small, concentrated spots between 50 and 100 μm (Figure 3b).
Nanomanipulator Matrix Spotting
Following analyte deposition, the matrix filled emitter was positioned immediately above the spot, and a sub-microliter amount was expelled with one pulse of the pressure injector. Matrix spots were able to be created as small as 57 μm, and proved to be tunable by increasing injection time using the pressure injector. To enable sufficient analysis time with extraction, the matrix spots created for this work were on average 485 ± 95 μm in size (Figure 3c). DHB was selected as a matrix here for its well documented and favorable characteristics for ionization of lipids, including its ionization efficiency at 337 nm, high achievable sensitivity, and lack of matrix interference in the m/z range for TAGs (m/z 750–1000) [38]. DHB at 20 mg/mL was used as the matrix concentration for MALDI-DOMS, which provided sufficient material to produce a mass spectrometry image (MSI) of the entire spot and allow a second pass to conduct MS/MS experiments without recoating the analyte with matrix. DHB, as noted previously [39, 40], is notorious for forming large (>50 μm) crystals during recrystallization from solvent. This can be especially problematic for tissue imaging mass spectrometry (IMS), where very small matrix crystal size is directly correlated with achievable spatial resolution of MS images and required to minimize delocalization of signal [40]. For MALDI-DOMS, as the analyte has already been dissolved in the extraction solvent and co-crystalized with the matrix, thus further delocalization of signal from large crystal size was inconsequential.
MALDI-DOMS MSI
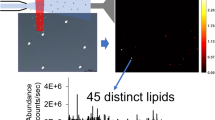
Figure 4a shows the MSI image of the total ion count of an extracted and deposited lipid droplet coated with matrix. After analysis, the MS image can be queried for the localization of a selected compound, and a heat map will be generated for the specified target. Here, the spot in Figure 4a was searched for the exact mass of TAG 50:2 [M + Na]+, at m/z 853.762 ± 0.003, which resulted in the image shown in Figure 4b. A total of 87 data points, and corresponding spectra, were obtained from this image. Furthermore, once the x and y coordinates of an analyte are known, location of the same coordinates can be re-analyzed for additional experiments. This is a significant improvement to the NSI-DOMS technique, where the analyte is quickly expelled from the emitter tip, ~30 s, limiting the potential exploration for MS/MS experiments. Using the MALDI-DOMS technique, we were able to analyze the same x and y coordinates up to four additional times after the initial full MS scan of the MSI. As proof of concept, we re-analyzed the spots in Figure 4b for MS/MS experiments. In 3T3-L1 adipocytes, TAG 50:2 can have a multitude of acyl compositions, such as (16:0/16:1/18:1), (16:0/16:0/18:2), or (16:1/16:1/18:0). MS/MS analysis is needed for proper assignment as the example molecular species have identical masses. We selected m/z 853.72 for collision-induced dissociation (CID) MS/MS experimentation, Figure 4c. Coupling the MALDI-DOMS technique with the high resolution and MS/MS capabilities of the instrument enabled ample analysis time and determination of the acyl composition of TAG 50:2. The resulting fragments (Figure 4d) indicated exact mass fatty acid losses of parent TAG composition (16:0/16:1/18:1).
(a) Total ion count of the co-deposited organelle content and matrix spots. (b) Selected TAG [50:2 + Na]+ at m/z 853.72 ± 0.003. (c) Fifty carbon acyl spectrum obtained from total ion count, with red box indicating TAG selected for MS/MS analysis. (d) Resulting spectrum from CID of m/z 853.7283 with fatty acid losses highlighted. One hundred μm scale bars
MALDI-DOMS Versus NSI-DOMS Data Analysis
MALDI-DOMS and NSI-DOMS techniques were used for lipid droplet extractions and the corresponding spectra of those lipid droplets are displayed. Figure 5 a, b illustrate the relative distribution of spectra from two separate lipid droplets, and 5c shows mol % comparison of the major TAG species from all extractions. The resolution of the Orbitrap provides clear baseline resolved and accurate mass ion signals defining the TAG profile obtained from the extractions. It should be noted, to explain the slight disparity in mass range, that the MALDI-DOMS spectra are adducted with sodium and the NSI-DOMS spectra are adducted with ammonium. The differences in the composition of TAG molecular species as shown in Figure 5c are illustrative to the heterogeneity of individual organelles and complementary to previous work [5, 14]. This 5%–10% difference in composition is in stark contrast to the ~1% RSD in the reproducibility of the standard (Figure 2). To further show the similarities in relative distribution of TAG molecular species obtained from the two ionization methods in similar sized lipid droplets, a close-up of the 48 carbon TAG region (Figure 6a, b, and c, d, respectively) is presented. The high, 100,000 resolution (at m/z 400), of MALDI-DOMS using an Orbitrap mass analyzer is clearly able to resolve the 48 carbon TAG species, and provides accurate mass (m/z ± 0.003) information regarding these molecular species. NSI-DOMS using a linear ion trap mass analyzer was able to provide compositional data regarding the 48 carbon TAGs; however, with the lower resolution, accurate mass is not able to be determined and limited mass analysis time with sample did not allow for MS/MS analysis.
Triacylglycerol molecular species profiles obtained from microextractions of individual lipid droplets using the described method. Spectrum (a) displays the profile obtained from the co-deposited extraction and matrix technique from the MALDI-LTQ-Orbitrap [M + Na]+. Spectrum (b) displays the TAG profile obtained from NSI [M + NH4]+ on a linear ion trap. Peak labels indicate the total number of carbons in the acyl chains followed by the total number of double bonds. (c) Mol% comparison of the major TAG species from all extractions
(a)-(b) Bright field image of lipid droplet microextraction and corresponding MALDI-DOMS mass spectrum of the 48 carbon TAG molecular species. TAG species were detected as [M + Na]+. (c)-(d) Bright field image of lipid droplet microextraction and corresponding NSI-DOMS mass spectrum of the 48 carbon TAG molecular. TAG species were detected as [M + NH4]+. Bright field images taken using 40x magnification. Scale bars represent 50 μm
Conclusions
Commonly used single cell and organelle extractions often involve lengthy chromatographic separations, purification steps, digestions, and/or harsh chemical treatments in which spatial context of the cells and organelles is lost during the extraction process [12, 13, 26, 33]. The precise targeting capability of the nanomanipulator avoids these issues by discriminately microextracting organelles, significantly reducing sample preparation time (<30 min for extraction and analysis), and leaving the remaining cell culture architecture intact. This attribute of the nanomanipulator is highly applicable for clinical diagnostics, where patient samples and cultures can be limited because the practitioner is only able to biopsy or remove small amounts of tissue. In plated cell cultures, the number of attainable microextractions using the nanomanipulation platform is only limited to the number of cells and how long the investigator is able to maintain the culture.
The results presented here validate this technique for single organelle analysis for cells in vitro. This allows for subsequent subcellular studies to probe cellular function and progression of diseases because the integrity of the remaining cell culture is uncompromised. Lipid droplets, because of their size, were targeted as a proof-of-concept method. Targeting other organelles would require adjustment of tip diameter and tailored solvent. The introduction of this technique is intended as a rapid platform of analyzing individual organelles selectively that provides more time with the extracted analyte than typical NSI and also leaves the remainder of the cell sample intact. This allows for additional analysis on following days, which would be especially useful for progressive diseases. Additional experiments will need to be performed to optimize this technique, such as determining ideal spot size, alternative matrices, and optimal laser energies.
References
Trouillon, R., Passarelli, M.K., Wang, J., Kurczy, M.E., Ewing, A.G.: Chemical analysis of single cells. Anal. Chem. 85, 522–542 (2013)
Jeffries, G.D.M., Edgar, J.S., Zhao, Y., Shelby, J.P., Fong, C., Chiu, D.T.: Using polarization-shaped optical vortex traps for single-cell nanosurgery. Nano Lett. 7, 415–420 (2007)
Oikawa, A., Saito, K.: Metabolite analyses of single cells. Plant J. 70, 30–38 (2012)
Zenobi, R.: Single-cell metabolomics: analytical and biological perspectives. Science 342, 1201 (2013)
Horn, P.J., Ledbetter, N.R., James, C.N., Hoffman, W.D., Case, C.R., Verbeck, G.F., Chapman, K.D.: Visualization of lipid droplet composition by direct organelle mass spectrometry. J. Biol. Chem. 286, 3298–3306 (2011)
Borland, L.M., Kottegoda, S., Phillips, K.S., Allbritton, N.L.: Chemical analysis of single cells. Annu. Rev. Anal. Chem. 1, 191–227 (2008)
Hebbar, S., Schulz, W.D., Sauer, U., Schwudke, D.: Laser capture microdissection coupled with on-column extraction LC-MSn enables lipidomics of fluorescently labeled drosophila neurons. Anal. Chem. 86, 5345–5352 (2014)
Espina, V., Wulfkuhle, J.D., Calvert, V.S., Van Meter, A., Zhou, W., Coukos, G., Geho, D.H., Petricoin III, E.F., Liotta, L.A.: Laser-capture microdissection. Nat. Protoc. 1, 586–603 (2006)
Ellis, S.R., Ferris, C.J., Gilmore, K.J., Mitchell, T.W., Blanksby, S.J., in het Panhuis, M.: Direct lipid profiling of single cells from inkjet printed microarrays. Anal. Chem. 84, 9679–9683 (2012)
Clemons, K., Wiley, R., Waverka, K., Fox, J., Dziekonski, E., Verbeck, G.F.: Direct analyte-probed nanoextraction coupled to nanospray ionization-mass spectrometry of drug residues from latent fingerprints. J. Forensic Sci. 58, 875–880 (2013)
Clemons, K., Dake, J., Sisco, E., Verbeck, G.F.: Trace analysis of energetic materials via direct analyte-probed nanoextraction coupled to direct analysis in real time mass spectrometry. Forensic Sci. Int. 231, 98–101 (2013)
Clemons, K., Nnaji, C., Verbeck, G.F.: Overcoming selectivity and sensitivity issues of direct inject electrospray mass spectrometry via DAPNe-NSI-MS. J. Am. Soc. Mass Spectrom. 25, 705–711 (2014)
Ledbetter, N.L., Walton, B.L., Davila, P., Hoffmann, W.D., Ernest, R.N., Verbeck, G.F.I.V.: Nanomanipulation-coupled nanospray mass spectrometry applied to the extraction and analysis of trace analytes found on fibers. J. Forensic Sci. 55, 1218–1221 (2010)
Phelps, M., Hamilton, J., Verbeck, G.F.: Nanomanipulation-coupled nanospray mass spectrometry as an approach for single cell analysis. Rev. Sci. Instrum. 85, 124101/1–124101/5 (2014)
Horn, P.J., Joshi, U., Behrendt, A.K., Chapman, K.D., Verbeck, G.F.: On-stage liquid-phase lipid microextraction coupled to nanospray mass spectrometry for detailed, nano-scale lipid analysis. Rapid Commun. Mass Spectrom. 26, 957–962 (2012)
Gibson, G.T.T., Mugo, S.M., Oleschuk, R.D.: Nanoelectrospray emitters: trends and perspective. Mass Spectrom. Rev. 28, 918–936 (2009)
Huynh, V., Joshi, U., Leveille, J.M., Golden, T.D., Verbeck, G.F.: Nanomanipulation-coupled to nanospray mass spectrometry applied to document and ink analysis. Forensic Sci. Int. 242, 150–156 (2014)
Phelps, M.S., Verbeck, G.F.: A lipidomics demonstration of the importance of single cell analysis. Anal. Methods 7, 3668–3670 (2015)
Goto-Inoue, N., Hayasaka, T., Zaima, N., Setou, M.: Imaging mass spectrometry for lipidomics. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 1811, 961–969 (2011)
Burnum, K.E., Frappier, S.L., Caprioli, R.M.: Matrix-assisted laser desorption/ionization imaging mass spectrometry for the investigation of proteins and peptides. Annu. Rev. Anal. Chem. 1, 689–705 (2008)
Svatos, A.: Mass spectrometric imaging of small molecules. Trends Biotechnol. 28, 425–434 (2010)
Zhang, Z., Kuang, J., Li, L.: Liquid chromatography-matrix-assisted laser desorption/ionization mass spectrometric imaging with sprayed matrix for improved sensitivity, reproducibility and quantitation. Analyst 138, 6600–6606 (2013)
Boggio, K.J., Obasuyi, E., Sugino, K., Nelson, S.B., Agar, N.Y.R., Agar, J.N.: Recent advances in single-cell MALDI mass spectrometry imaging and potential clinical impact. Expert Rev. Proteomics 8, 591–604 (2011)
Strupat, K., Kovtoun, V., Bui, H., Viner, R., Stafford, G., Horning, S.: MALDI produced ions inspected with a linear ion trap-Orbitrap hybrid mass analyzer. J. Am. Soc. Mass Spectrom. 20, 1451–1463 (2009)
Garden, R.W., Shippy, S.A., Li, L., Moroz, T.P., Sweedler, J.V.: Proteolytic processing of the Aplysia egg-laying hormone prohormone. Proc. Natl. Acad. Sci. U. S. A. 95, 3972–3977 (1998)
Whittal, R.M., Keller, B.O., Li, L.: Nanoliter chemistry combined with mass spectrometry for peptide mapping of proteins from single mammalian cell lysates. Anal. Chem. 70, 5344–5347 (1998)
Jimenez, C.R., Li, K.W., Dreisewerd, K., Spijker, S., Kingston, R., Bateman, R.H., Burlingame, A.L., Smit, A.B.., van Minnen, J., Geraerts, W.P.M.: Direct mass spectrometric peptide profiling and sequencing of single neurons reveals differential peptide patterns in a small neuronal network. Biochemistry 37, 2070–2076 (1998)
Lanni, E.J., Rubakhin, S.S., Sweedler, J.V.: Mass spectrometry imaging and profiling of single cells. J. Proteomics 75, 5036–5051 (2012)
Rubakhin, S.S., Lanni, E.J., Sweedler, J.V.: Progress toward single cell metabolomics. Curr. Opin. Biotechnol. 24, 95–104 (2013)
Lorenzo, T.M., Mizuno, H., Tsuyama, N., Harada, T., Masujima, T.: In situ molecular analysis of plant tissues by live single-cell mass spectrometry. Anal. Chem. 84, 5221–5228 (2012)
Aerts, J.T., Louis, K.R., Crandall, S.R., Govindaiah, G., Cox, C.L., Sweedler, J.V.: Patch clamp electrophysiology and capillary electrophoresis-mass spectrometry metabolomics for single cell characterization. Anal. Chem. 86, 3203–3208 (2014)
Masujima, T.: Live single-cell mass spectrometry. Anal. Sci. 25, 953–960 (2009)
Rubakhin, S.S., Garden, R.W., Fuller, R.R., Sweedler, J.V.: Measuring the peptides in individual organelles with mass spectrometry. Nat. Biotechnol. 18, 172–175 (2000)
Chandra, S., Lorey, D.R.I.I.: SIMS ion microscopy in cancer research: single cell isotopic imaging for chemical composition, cytotoxicity and cell cycle recognition. Cell. Mol. Biol. 47, 503–518 (2001)
Lysy, P.A., Smets, F., Sibille, C., Najimi, M., Sokal, E.M.: Human skin fibroblasts: from mesodermal to hepatocyte-like differentiation. Hepatology 46, 1574–1585 (2007)
Madsen, L., Petersen, R.K., Kristiansen, K.: Regulation of adipocyte differentiation and function by polyunsaturated fatty acids. Biochim. Biophys. Acta Mol. Basis Dis. 1740, 266–286 (2005)
Ntambi, J.M., Kim, Y.-C.: Adipocyte differentiation and gene expression. J. Nutr. 130, 3122S–3126S (2000)
Schiller, J., Süß, R., Arnhold, J., Fuchs, B., Leßig, J., Müller, M., Petković, M., Spalteholz, H., Zschörnig, O., Arnold, K.: Matrix-assisted laser desorption and ionization time-of-flight (MALDI-TOF) mass spectrometry in lipid and phospholipid research. Prog. Lipid Res. 43, 449–488 (2004)
Fuchs, B., Süß, R., Schiller, J.: An update of MALDI-TOF mass spectrometry in lipid research. Prog. Lipid Res. 49, 450–475 (2010)
Hankin, J.A., Barkley, R.M., Murphy, R.C.: Sublimation as a method of matrix application for mass spectrometric imaging. J. Am. Soc. Mass Spectrom. 18, 1646–1652 (2007)
Acknowledgments
This work was supported by the CPRIT High-Impact Research Award (RFA R-13-HIHR-1) to G.F.V., and in part by a grant to K.D.C. from the US Department of Energy, Office of Science, Basic Energy Sciences program (DE-FG02-05ER15647). The authors thank Roberto Aguilar for his work in preparing computer-aided design (CAD) images of the nanomanipulator workstation.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Online Resource 1
Video displaying the DOMS extraction process of a single lipid droplet performed by the nanomanipulator. (MPG 1668 kb)
Online Resource 2
Video showing the co-depositon process used for MALDI-DOMS. The first nanospray emitter (top) is shown pulse-spotting the extracted organelle content. The second emitter (left) is shown depositing matrix solution atop the original spot. (MPG 2271 kb)
Rights and permissions
About this article
Cite this article
Phelps, M.S., Sturtevant, D., Chapman, K.D. et al. Nanomanipulation-Coupled Matrix-Assisted Laser Desorption/ Ionization-Direct Organelle Mass Spectrometry: A Technique for the Detailed Analysis of Single Organelles. J. Am. Soc. Mass Spectrom. 27, 187–193 (2016). https://doi.org/10.1007/s13361-015-1232-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-015-1232-9