Abstract
Mass spectrometric de-novo sequencing was applied to review the amino acid sequence of a commercially available recombinant protein G´ with great scientific and economic importance. Substantial deviations to the published amino acid sequence (Uniprot Q54181) were found by the presence of 46 additional amino acids at the N-terminus, including a so-called “His-tag” as well as an N-terminal partial α-N-gluconoylation and α-N-phosphogluconoylation, respectively. The unexpected amino acid sequence of the commercial protein G′ comprised 241 amino acids and resulted in a molecular mass of 25,998.9 ± 0.2 Da for the unmodified protein. Due to the higher mass that is caused by its extended amino acid sequence compared with the original protein G′ (185 amino acids), we named this protein “protein G′e.” By means of mass spectrometric peptide mapping, the suggested amino acid sequence, as well as the N-terminal partial α-N-gluconoylations, was confirmed with 100% sequence coverage. After the protein G′e sequence was determined, we were able to determine the expression vector pET-28b from Novagen with the Xho I restriction enzyme cleavage site as the best option that was used for cloning and expressing the recombinant protein G′e in E. coli. A dissociation constant (K d ) value of 9.4 nM for protein G′e was determined thermophoretically, showing that the N-terminal flanking sequence extension did not cause significant changes in the binding affinity to immunoglobulins.
ᅟ
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Since DNA sequencing was introduced in the mid-seventies of the last century, it gained great importance in suggesting amino acid sequences of proteins by simple translation of the gene sequence [1]. However, significant possibilities of amino acid sequence aberrations due to mutations, amino acid substitutions in (recombinant) proteins (e.g., by wobbling, [2]), or by altering the expression system, are inherent to this DNA-based protein sequence determination approach [3, 4]. Unexpected post-translational modifications (PTMs) are not accessible.
Continuously growing possibilities of mass spectrometry-based fragmentation techniques, such as collisional induced dissociation (CID) and electron capture dissociation (ECD), enormously facilitate direct sequence determination of even fairly large intact proteins by so-called “top-down” protein sequencing [5].
Consequently, this mass spectrometry-driven amino acid sequencing approach opens the opportunity to revise DNA-derived sequence information of many proteins [6, 7]. The importance of these MS-based sequencing avenues for scientific projects has been emphasized by the fact that deviations in previously annotated amino acid sequences of several recombinant proteins have been reported [8–10]. Here we apply this mass spectrometry-driven amino acid sequencing approach to protein G′, a commercially available protein with great scientific and economic importance that is available from many companies around the world.
Protein G was discovered as a cell-surface protein of different Streptococcus species in 1973 [11], and first amino acid sequences were reported in the mid-eighties [12, 13]. Its astounding binding properties to mammalian immunoglobulin G (IgG) fostered extensive research on functional optimization up to the mid-nineties [14–19]. Depending on the streptococcal strain, protein G contains, in addition to three domains for IgG binding, two or three domains that bind to mammalian serum albumin [20, 21]. Initial difficulties in purification of protein G directly from the streptococcal cell wall were overcome after the DNA sequence of its encoding gene was successfully overexpressed in E. coli [12, 22]. Later, truncated genes (e.g., from the Streptococcus strain G 148) that encoded just for the three IgG binding domains were cloned and expressed in E. coli. The shorter protein was named protein G′ [23] to differentiate it from the full-length protein G. Owing to its extraordinary high binding affinity to immunoglobulins, protein G′ is now widely used in many immunologically and biotechnologically applied techniques world-wide. When coupled to a chromatography resin, protein G′ has become an indispensable workhorse for affinity purification of antibodies and of Ig-tagged recombinant proteins [24]. Versatile applications of protein G′ have been reported numerously (reviewed in [25, 26]), from which only a few shall be mentioned: isolation of IgG fractions from patient samples; immuno-precipitation [27, 28]; depletion of IgG from biological samples [29, 30]; Western blot analysis [31]; affinity membrane chromatography [32]; peptide immunoaffinity enrichment using protein-G′ coated magnetic beads [33]; development of protein G′-coupled receptors [34]; and generation of immunosensors [35].
For studying the principles of function and the dynamics of protein G′-binding to IgG, knowledge of its structure is a prerequisite. Hence, the first piece of information, when conducting a study on protein—protein interactions, is to collect the amino acid sequences of both interaction partners. For protein G′ this requirement sounds trivial, as recombinant protein G′-containing products can be found in catalogs of almost every supplier in the biotechnological field, including Sigma-Aldrich, Merck-Millipore, Thermo-Scientific, Life-Technologies, and Biocat, to name just a few. According to the product information provided by the suppliers, the commercial protein G′ carries three IgG binding domains, which calculate to a molecular mass of ca. 20 kDa. Yet, on sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), protein G′ shows an apparent molecular weight of ca. 35 kDa [12]. Strikingly, despite the huge sales market for protein G′, information about the amino acid sequence of the commercial products is poor. Vendors of recombinant protein G′ are rarely able to provide the amino acid sequence of their product. Upon request, customers are referred to the literature from the 1980s and 1990s. Although the amino-acid sequence that is given in the respective reports stands in agreement with the molecular mass of 20 kDa for protein G′ [23], the mass of the commercial product does not. Applying mass spectrometric analysis to the product in our hands, we found a mass increase of 6 kDa for which no explanation was retrievable. Information about the existence of a His-tag and sometimes of biotinylation did not explain the mass difference. Unfortunately, the aberrant SDS-PAGE migration behavior of protein G′ prevents easy discovery of any size-related irregularities in the protein under study.
Thus, to study the binding properties and possible influences on these interactions of the mutual additional parts in protein G′, we first had to determine the amino acid sequence of the commercially available product, from here on referred to as “protein G prime e (protein G′e).” We employed mass spectrometry-based top-down “de-novo” sequencing, assisted by bottom-up approaches, for elucidating its amino acid sequence and potential modifications. The newly determined amino acid sequence of protein G′e was confirmed by mass spectrometric peptide mapping. Finally, we assessed the dissociation constant of protein G′e towards IgG binding by microscale thermophoresis.
Materials and Methods
SDS PAGE Analysis of Intact Protein G′e
Protein G′e was obtained as lyophilized powder from Sigma (catalog no. P4689-5MG; lot no. SLBB8536V). A stock solution (2 μg/μL) of protein G'e was prepared by dissolving 500 μg of the protein in 250 μL of 100 mM ammonium bicarbonate, pH 8. Next, the stock solution was diluted with 100 mM ammonium bicarbonate, pH 8, to obtain a working solution with a final protein G′e concentration of 1 μg/μL. One μL of this protein G′e working solution was mixed with 9 μL of water and 2.5 μL of SDS sample buffer (312.5 mM tris-(hydroxymethyl) aminomethane (TRIS), 10% SDS, 50% glycerol, 325 mM dithiothreitol (DTT) and 6 mM bromophenol blue). This mixture was loaded directly onto a NuPAGE 10% Bis-Tris gel (Invitrogen, Karlsruhe, Germany). The protein mass marker (Broad Range, New England BioLabs, Frankfurt/Main, Germany) was used to determine the apparent molecular mass of the loaded sample. The gel was placed into an electrophoresis chamber, and power was applied for 1 h at 200 V; 3-(N-morpholino) propanesulfonic acid (MOPS) buffer (25 mM MOPS, 25 mM TRIS, 3.5 mM SDS, and 1 mM ethylenediamine tetra-acetic acid) were used as running buffer. Afterwards, the gel was removed from the plates, and the proteins were fixed in the gel by bathing it for 1 h at room temperature in 50 mL of fixation solution (50% ethanol, 10% acetic acid). Proteins were stained overnight at room temperature with 50 mL colloidal Coomassie brilliant blue G250 (CBB G250) solution that contains 2.3% phosphoric acid (85%), 10% ethanol, 5% aluminum sulfate 14–18 hydrate, and 0.02% CBB G250. Then, the gel was washed with 50 mL of destaining solution [2.3% phosphoric acid (85%), 10% ethanol] twice for 1 h, each, at room temperature. Stained gels were immediately scanned with the Umax Mirage II Scanner (Umax Data Systems, Willich, Germany) [36].
Desalting of Intact Protein G′e
First, reversed phase (RP)-packed tip material (ZipTip C4 tips; Millipore, Billerica, MA, USA) was reconstituted using 50% ACN, pH 5.8, and an equilibration solution (0.1% TFA, pH 1.7), respectively, by aspirating and dispensing 10 μL of each solution twice. Next, 5 μL of protein G′e working solution (see above) was mixed with 5 μL of equilibration solution and loaded onto the RP-packed tip (Zip Tip) material by aspirating and dispensing 10 times. Next, washing was performed by aspirating and dispensing twice using 10 μL of equilibration solution. Then, protein G′e was eluted with 5 μL of 80% ACN, 0.1% TFA, pH 1.7, by passing it through the RP-packed tip (ZipTip) material 10 times. The resulting concentration of protein G′e was 0.3 μg/μL. The concentration was determined using the Bio-Rad Protein Assay (Bio-Rad, Munich, Germany) [37, 38].
Nano-ESI MS Analysis of Intact Protein G′e
Protein G′e (150 μg) was dissolved in 150 μL of 2% aqueous acetic acid:MeOH (95:5, v/v), pH 2.5, to obtain a final protein concentration of 1 μg/μL. 5 μL of this solution was loaded into an EconoTip emitter (ECONO10; New Objective Inc., Woburn, MA, USA) using a microloader pipette tip (Eppendorf, Hamburg, Germany). Mass spectra were acquired in the positive-ion mode using a Waters electrospray ionization (ESI) Q-ToF II mass spectrometer (Waters MS-Technologies, Manchester, UK), setting the mass window to m/z 100–4000 [39]. The following experimental parameters were used for all measurements: capillary voltage, 1.5 kV; extractor cone, 3 V; radio frequency (rf) lens, 1.2 V; source temperature, 60°C; nitrogen counter flow gas, 50 L/h; scan rate, 7 s/scan; digitization rate, 4 GHz; microchannel plate detector voltage, 1950 V. Sample cone voltage settings were changed between 60 V and 160 V. Data acquisition and processing was performed with the MassLynx software ver. 4.0 (Waters MS-Technologies). External calibration was performed with 1% phosphoric acid dissolved in a trifluoroethanol/water solution (50:50, v/v) [40, 41].
Top-Down Protein Sequencing of Intact Protein G′e by ESI-ECD-FT-ICR-MS
A syringe pump (Harvard PHD Ultra syringe pump; Instech Laboratories, Inc, Plymouth Meeting, PA, USA) was used to infuse protein G′e (0.26 μg/μL dissolved in 0.1% FA in 60% ACN) at a flow rate of 200 nL/min for nano-electrospray ionization. The nano-ESI tips were prepared in-house from silica capillary tubing of 360 μm outer and 150 μm inner diameters (Polymicro Technologies, Phoenix, AZ, USA) by using a laser-based micropipette puller P-2000 (Sutter Instrument Co., Novato, CA, USA). ECD-based Fourier transform-ion cyclotron resonance (FT ICR) top-down sequencing was performed on a Bruker SolariX 12 T Fourier transform-ion cyclotron resonance (FT-ICR) mass spectrometer (Bruker Daltonics, Bremen, Germany) [42]. The following instrumental settings were used: nano-ESI voltage, 0.9 kV; drying gas temperature, 180°C; drying gas flow, 1.0 L/min. The Bruker control software does not provide a direct readout of the setting delay between ion trapping and electron irradiation. The delay is estimated at tens of microseconds. The 26+ ion signal was chosen for isolation in the quadrupole region prior to transfer to the ICR trap. A collision voltage of 2 V was applied for activation of the precursor ions prior to ECD. The ECD cathode heater current was 1.6 A, the bias was −0.4 eV, and duration of the electron beam was 0.1 ms. The FT ICR mass spectrometer was externally calibrated with ubiquitin. Four thousand scans of 1 M data point spectrum were averaged. The acquired mass spectra were further processed and analyzed with the Data Analysis software (ver. 4.0. SP 5; Bruker Daltonics). Ion peaks were labeled by using the SNAP peak picking algorithm. Signal-to-noise threshold was set to 2 and the quality factor threshold to 0.6. Fragment ion mass assignment was performed using the BioTools 3.0 software (Bruker Daltonics), resulting in a MS/MS search error tolerance of below 25 ppm upon external calibration with ECD fragments of ubiquitin. The presumed amino acid sequence was edited and C″-type and Z′-type ions were automatically annotated and manually checked [43].
MALDI-ToF MS Analysis of Intact Protein G′e
A volume of 0.8 μL of desalted protein G′e (0.24 μg) was deposited on an AnchorChip 400/384 target plate (Bruker Daltonics) and mixed directly on the target with 2 μL of 2,5-dihydroxybenzoic acid solution (DHB; LaserBio Labs, Sophia-Antopolis, France; 5 μg/μL DHB in water/ACN/TFA; 49.9/50/0.1, v/v/v). MALDI spectra were acquired by using a Reflex III MALDI ToF mass spectrometer (Bruker Daltonics), equipped with a SCOUT source, in positive-ion linear mode, setting the acceleration voltage to 20 kV. The mass window was set to 0.7–41.3 kDa. A nitrogen laser (wavelength 337 nm, pulse width 3–5 ns) was used for desorption. Around 600 laser shots per spectrum were summed and the accumulated spectra were analyzed with the FlexAnalysis 2.4 software (Bruker Daltonics). Spectra were externally calibrated using the protein calibration standard I (Bruker Daltonics) [44, 45].
Protein Sequencing by MALDI-ISD-MS
A protein G′e stock solution (see above; 0.5 μL) was deposited on an AnchorChip 400/384 target plate (Bruker Daltonics). After complete solvent evaporation, 0.8 μL of sinapinic acid [LaserBio Labs, France; 10 mg/mL in EtOH:acetone (67:33, v/v)] was added. After drying, the matrix solution addition (0.8 μL, each) was repeated twice. Next, the dried sample–matrix mixture was washed with 5 μL of 1% TFA solution. Matrix-assisted laser desorption/ionization (MALDI)-in source decay (ISD) mass spectra were acquired on a Reflex III MALDI ToF mass spectrometer (Bruker Daltonics) equipped with a SCOUT source. Each single MALDI-ISD mass spectrum was acquired from 5000 summed laser shots in the positive-ion mode over an m/z range 1000–10,000. Laser power was set at 80%–90% to increase fragmentation. The acceleration voltage was set to 20 kV, and reflector voltage was 23 kV. Further data processing and analysis was performed using FlexAnalysis 4.2 and BioTools 3.0 software (Bruker Daltonics). For ISD analysis, spectra were permanently assigned as “ISD-type.” Monoisotopic masses were labeled, and ion-signal assignment was performed manually by subtracting m/z values of the neighboring signals, which correspond to loss of one amino acid. Next, deduced amino acid sequence was loaded into the sequence editor of BioTools and C″- and Y″-type ions were automatically annotated to verify the manual assignment. With a mass tolerance of 0.7 Da, accurate assignment was possible for the ion signals with higher m/z values above m/z 4000 and at the same time the assignment was accurate enough to correctly assign ion signals below m/z 4000 [46].
In-Solution Digestion of Protein G′e with Trypsin
Protein G′e working solution (25 μL; see above), was subjected to in-solution digestion with trypsin (Promega, Madison, WI, USA, reconstituted according to the manufacturer’s protocol) by using an enzyme to substrate ratio of 1:100 (w/w). Digestion was performed at room temperature overnight. To stop digestion, the protein–enzyme mixture was frozen and kept at −20°C [47].
Asp-N In-Solution Digestion of Protein G′e
Protein G′e working solution (25 μL; see above), was subjected to in-solution digestion with Asp-N (Roche, Mannheim, Germany; reconstituted according to the manufacturer’s protocol) by using an enzyme to substrate ratio of 1:50 (w/w). Digestion was performed at room temperature overnight. To stop the digestion, the protein–enzyme mixture was frozen and kept at −20°C [48].
Desalting of Peptides from In-Solution Digestions
A volume of 5 μL of peptide solution derived from tryptic or Asp-N in-solution digestion of protein G′e (see above) was desalted with RP-packed tip (ZipTip C18 tips; Millipore, Billerica, MA, USA). The RP-packed tip (ZipTip) material was reconstituted with 50% ACN, pH 5.8, and with an equilibration solution (0.1% TFA, pH 1.7) by aspirating and dispensing 10 μL of each solution twice. Next, 5 μL of the digest were mixed with 5 μL equilibration solution, and from this solution peptides were loaded onto the RP-packed tip (ZipTip) material by aspirating and dispensing all the volume 10 times. Afterwards, salts were removed with twice 10 μL of 0.1% TFA, pH 1.7. Peptides were eluted with 5 μL of 80% ACN, 0.1% TFA, pH 1.7, by passing it through the RP-packed tip (ZipTip) device 10 times [49].
MALDI-ToF-MS Peptide Mapping
A volume of 0.8 μL of the protein G′e peptide mixture after desalting was prepared onto an AnchorChip 400/384 target plate (Bruker Daltonics) with 2 μL of DHB (LaserBio Labs) matrix solution (5 μg/μL DHB in water/ACN/TFA, 49.9/50/0.1 v/v/v). The preparation was allowed to dry, and the target plate was introduced into the SCOUT source of the Reflex III MALDI ToF mass spectrometer (Bruker Daltonics). Spectra of protonated peptides (summing up about 600 laser shots) were acquired either in reflector mode (mass window m/z 400–5000), or in linear mode (mass window m/z 2250–40,600). Acceleration and reflector voltages were 20 and 23 kV, respectively. Spectra were externally calibrated by using the peptide calibration standard (reflector mode) and the protein calibration standard (linear mode) from Bruker Daltonics and recalibrated internally using peptide ion signals derived from trypsin autoproteolysis. Mass spectra were further processed and analyzed using the FlexAnalysis 4.2 and BioTools 3.0 software (Bruker Daltonics). Peptide ion signals were assigned and interpreted manually comparing experimental m/z values with a peak list obtained from the theoretical digest of the presumed amino acid sequence of protein G′e, using the GPMAW ver. 9.1 software (Lighthouse Data, Odense, Denmark) [50, 51]. MS error tolerance for peptide mass fingerprinting was between 20 and 30 ppm.
MALDI-QIT-ToF MS/MS Fragmentation
The protein G′e peptide mixture after desalting (0.8 μL) was prepared on an AnchorChip 400/384 target plate (Bruker Daltonics). DHB (2 μL) (LaserBio Labs) matrix solution (5 μg/μL DHB in water/ACN/TFA, 49.9/50/0.1 v/v/v) was added. After drying, product ion (MS/MS) spectra were acquired on an Axima MALDI quadrupole ion trap (QIT) time of flight (ToF) mass spectrometer (Shimadzu Biotech, Manchester, UK) in the positive-ion mode by utilizing a 337 nm nitrogen laser and a three-dimensional quadrupole ion trap supplied with a pulsed helium flow gas for cooling and argon gas to cause collisionally induced dissociation [52]. Spectra were calibrated externally with a manually prepared peptide mixture composed of bradykinin (1–7) [M + H]+ 757.39, angiotensin II [M + H]+ 1046.53, angiotensin I [M + H]+ 1296.68, bombesin [M + H]+ 1619.81, N-acetyl renin substrate [M + H]+ 1800.93, ACTH (1–17) [M + H]+ 2093.08, ACTH (18–39) [M + H]+ 2465.19, somatostatin [M + H]+ 3147.46, and insulin (oxidized beta chain) [M + H]+ 3494.64. For each spectrum, approximately 2500 profiles were summed. Spectra processing and analysis were performed by using the Launchpad software, ver. 2.7.1 (Shimadzu Biotech). Ion-signal assignment and sequence analysis was performed with the de-novo sequencing software SeqLab ver. 1.5 (Shimadzu Biotech), and signal assignments were verified manually [53].
Protein G′e Interaction Analysis with IgG
A solution of 100 μL of protein G′e (10 μM) dissolved in 100 mM ammonium acetate buffer, pH 6.9, was labeled with 100 μL of the red fluorescent dye NT-647 (reconstituted according to the manufacturer’s protocol) using the Monolith Protein Labeling Kit RED-NHS (NanoTemper Technologies, Munich, Germany). After labeling, free dye was removed via filtration through Gravity Flow Column B (NanoTemper Technologies), and purified labeled protein G′e was collected by adding 600 μL of a MicroScale Thermophoresis optimized buffer (containing 50 mM Tris (pH 7.6), 150 mM NaCl, 10 mM MgCl2, and 0.05% Tween-20) to the column [54]. Next, protein-to-dye ratio was determined spectroscopically. The number of fluorescent counts of labeled protein G′e was compared with a calibration curve of the dye alone and, from this, the approximate concentration of labeled protein G′e was found to be 20 nM. Protein G′e concentration was kept constant during the subsequent experiments. For affinity quantification, 16 samples of intravenous immunoglobulin (IVIg) (Omrix Biopharmaceuticals, Nes-Ziona, Israel) were prepared in 1:1 serial dilutions with the highest final concentration of 500 nM. Dilutions were performed by using a MicroScale Thermophoresis optimized buffer (see above) without Tween-20 [55]. Volumes of 10 μL of each serial dilutions of IgG were mixed with 10 μL of the fluorescently labeled protein G′e solution and incubated for approximately 10 min at room temperature. Next, each sample was loaded into one “MicroScale Thermophoresis Standard Treated Capillary” (NanoTemper Technologies) by means of capillary force. A dissociation constant (K d ) determination was performed with a Monolith NT.115 instrument using 20% MST power and 90% LED power. Laser-on time was 30 s and laser-off time was 5 s. As data output, the NanoTemper analysis software automatically plotted Fnorm values as a function of IgG concentration (titration curve with arbitrary intensity units) and calculated the K d value from the curve approximation by software-implemented algorithms [56].
Results
Molecular Mass Determination of Protein G′e
From the suggested amino acid sequence (Uniprot: Q54181), a molecular mass of 20,118.0 Da (average mass) was calculated for protein G′. SDS-PAGE analysis showed a single, well-stained band for protein G′e at an apparent molecular mass of ca. 35 kDa (Figure 1 right), indicating high purity and homogeneity. The anomalously high apparent molecular mass in SDS-PAGE stands in agreement with previous reports [23, 39].
Molecular mass analysis of protein G′e. SDS-PAGE analysis of protein G′e (right) shows an anomalous apparent molecular mass of ca. 35 kDa. Lane 1: protein mass markers. Lane 2: protein G′e (1.0 μg). Proteins were stained with colloidal Coomassie blue. Nano-ESI mass spectrum of protein G′e (bottom). Protein G′e (19.2 μM) was dissolved in 2% aqueous acetic acid: methanol (95:5, v/v), pH 2.4. Multiply protonated protein species are labeled. Two adjacent ion series (a and b) are found (see zoom view). For molecular mass assignment see Supplementary Table 1
NanoESI-MS analysis of protein G′e under acidic conditions showed a homogenous ion series of multiply charged ions with charge states from [M + 14H]14+ to [M + 23H]23+ in the m/z range between 1000 and 3000 with a maximum at the signal for the [M + 20H]20+ protein ion (Figure 1 bottom and Supplementary Table 1). Under denaturing conditions, narrow molecular ion signals were obtained that allowed differentiation of two closely spaced ion series (a and b). Surprisingly, the experimentally determined molecular masses (average masses) were 25,998.9 ± 0.2 Da (a ion series) and 26,177.2 ± 0.5 Da (b ion series), respectively, indicating the presence of two protein species. Yet, neither of the two experimentally determined molecular masses matched the calculated mass of protein G′, indicating that the commercial protein was not protein G′ but possessed a different amino-acid sequence. The heavier protein G′e presumably harbored a covalent modification with a mass of 178.3 Da. As neither the amino acid sequence database (Uniprot: Q54181) nor the provider (Sigma) was helpful to clarify the situation, we went on and determined the amino acid sequence “de-novo” by mass spectrometry.
Protein G′e “De-Novo” Sequence Determination
Our first “de-novo” mass spectrometric “top-down” sequencing attempt made use of the MALDI-ISD-ToF-MS method and employed a total of 38.5 pmol of protein G′e that was placed on the AnchorChip target and mixed with sinapinic acid as matrix. The initial survey by linear mode MALDI-ToF-MS presented a spectrum with strong signals corresponding to singly and doubly charged proteins (Supplementary Figure 1 and Supplementary Table 2), indicating an adequate quality of the preparation.
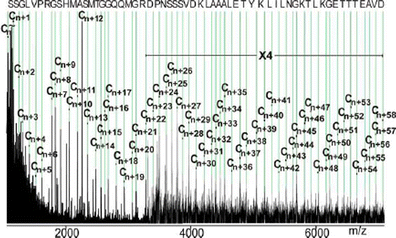
MALDI-ISD-ToF-MS top-down sequencing produced primarily C″-type fragment ions [46, 57] and best ISD fragmentation results were obtained using sinapinic acid matrix. Judging from the fragment ion with the lowest m/z value (C″n; m/z 1071.11), a short N-terminal partial sequence of 9 to 10 amino acids was left unobserved. The mass difference between the C″n ion and the C″n+1 ion (Δm 87.05) was indicative of a serine residue. In total, 59 amino acids from the N-terminus could be identified by reading the complete “C″-ion ladder” (Figure 2 and Supplementary Table 3). In two cases, larger distances between adjacent C″ ions were found than expected for single amino-acid residues, indicating the presence of a peptide bond N-terminal to a proline residue [58]. A gap of 196.24 mass units was found between the intense ion signals C″n+4 and C″n+6 (m/z 1415.38 and 1611.62). Subtracting the mass of a proline residue from the observed mass difference left the mass increment of a valine residue. In fact, at m/z 1515.41 a poorly resolved ion signal was found, confirming the “VP” dipeptide sequence. Similarly, the mass difference of 211.08 between ion signals C″n+22 at m/z 3284.40 and C″n+24 at m/z 3496.64 could be assigned to the dipeptide “DP.” Again, a low-intensity ion signal was observed at m/z 3399.46 and substantiated the assignment. Interestingly, starting from the C″n+48 ion signal, the next eleven C″ ion signals matched precisely with the first 11 amino acid residues from the IgG binding part of protein G′ (Uniprot: Q54181). Reading into the protein G′ sequence enabled us to place the newly identified partial sequence at the N-terminus of protein G′e and confirmed the presence of C″-type fragment ions in the spectrum. Note, when aligning our newly determined sequence with the full length protein G (Uniprot: P19909), the identical sequence part of both could be extended to 21 amino acids (starting from C″n+38).
Top-down MALDI-ISD mass spectrometric sequencing of protein G′e. C″-type ions are labeled (abbreviated as Cn ions to reduce complexity, where n is the number of the amino acid residues). A sequence of 59 amino acids from the N-terminal region is identified (de-novo sequencing) by reading the “C″ ion ladder”; mass tolerance was set to 0.7 Da; sinapinic acid was used as matrix. For molecular mass assignment see Supplementary Table 3
As the very N-terminal amino acid sequence (ca. 9 to 10 residues) was not yet determined, a bottom-up “de-novo” sequencing experiment was performed using a MALDI-QIT-ToF-MSn instrument. Upon in-solution tryptic digestion of protein G′e, the MALDI-ToF mass spectrum of the resulting peptides displayed six strong peptide ion signals of m/z 1535.71, 1768.93, 1909.01, 1946.98, 2465.10, and 3425.47 (Supplementary Figure 2 and Supplementary Table 4) together with approximately two dozen ion signals of rather low abundance. As none of the intense ion signals could be matched to the protein G′ sequence (Q54181) or to the full-length protein G sequence (P19909), they were assumed to belong to the N-terminal flanking amino acid sequence and subjected to mass spectrometric fragmentation.
The precursor ion with signal at m/z 1768.93 yielded in the amino acid sequence GSSHHHHHHSSGLVPR by MS/MS fragmentation (Figure 3a and Supplementary Table 5). The sequence of consecutively assembled six histidine residues proved the existence of a so-called “His-tag” at the N-terminus. This peptide also defined the very N-terminus of protein G′e and, starting with the SSGL-sequence, matched the suggested amino acid sequence obtained by MALDI-ISD-ToF-MS. Using a “de novo” sequencing software of the MALDI-QIT-ToF-MSn instrument, we were able to also deduce the amino acid sequence of the peptide of m/z 1946.99. The best match is the amino acid sequence [178]GSSHHHHHHSSGLVPR, the same sequence as the one for the peptide of m/z 1768.93, only with an extra mass of 178.3 Da (marked with [178]) at its N-terminus (Figure 3b; Supplementary Table 6). This mass increment of 178.3 Da corresponds to an N-terminal gluconoylation that was occasionally found in recombinant proteins that contain an N-terminal “GSS-His-tag” and were expressed in E. coli [59]. Note, this mass increment was already detected by ESI-MS analysis of the intact protein G′e.
Bottom-up MALDI-QIT-ToF MS/MS sequencing of protein G′e. (a) Fragment ion spectrum from precursor peptide with ion signal at m/z 1768.93. The amino acid sequence of the peptide is covered by Y″-type and B-type ions. (b) MALDI-QIT-ToF product-ion (MS/MS) spectrum of fragment ions from peptide ion of m/z 1946.99. The amino acid sequence of the peptide is covered by Y″-type and B-type ions. Presence of an N-terminal extra mass of 178 Da is shown. For molecular mass assignment see Supplementary Tables 5 and 6
MS/MS fragmentation of the precursor ion of m/z 1535.71 produced abundant B-type and Y″-type ions (Supplementary Figure 3; Supplementary Table 7) from which the sequence GSHMASMTGGQQMGR could be determined by using the aforementioned “de novo” sequencing software. This peptide sequence matched precisely with the partial sequence that was deduced by MALDI-ISD-ToF-MS to cover ions from C″n+8 to C″n+22. Similarly, the peptide ion of m/z at 1909.01 also produced B-type and Y″-type ion series. From them, two large peaks stood out at m/z 1107.75 and 1794.01. This stands in agreement with cleavage of D–P and D–K bonds, respectively (Supplementary Figure 4; Supplementary Table 8). Note, the determined partial sequence SVDKLAAALETY reads into the protein G sequence (P19909), again standing in agreement with the MALDI-ISD-ToF-MS top-down sequencing results.
By contrast, two peptide ions of m/z at 2465.10 and 3425.47 gave rise to just two abundant fragment ions instead of producing an extended fragment ion series. Most interestingly, both precursors yielded fragment ions with exactly the same m/z values at 1632.63 and 2318.94 (Supplementary Figure 5; Supplementary Tables 9 and 10). Obviously, most of the CID energy was consumed to cleave the peptide bonds at these two (predetermined) breaking points, likely at aspartic acid residues, and suppressed further fragmentation. Matching both precursor ion masses to the newly determined amino acid sequence by MALDI-ISD-ToF-MS aligned the ion signal at m/z 2465.10 to the partial sequence GSHMASMTGGQQMGRDPNSSSVDK and the ion signal at m/z 3425.47 to the partial sequence GSHMASMTGGQQMGRDPNSSSVDKLAAALETYK. The partial sequence of the precursor of m/z 3425.47 extends at the C-terminal end owing to a missed cleavage at a “K” residue that is located next to a “D.” Given that both peptides share the same N-terminal sequence, cleavage between dipeptides “DP” and “DK” produced the same B-type ions. The location of the “DP” dipeptide adjacent to the arginine residue also explains the missed cleavage at this residue.
Combining the newly determined N-terminal sequence information that was obtained from protein G′e by both MALDI-ISD-MS and MALDI QIT-ToF MS/MS with the amino acid sequence of protein G′ (Q54181) allowed us to assemble an amino acid sequence for the unmodified protein G′e. This sequence contained 241 amino acid residues from which a molecular mass of 25,999.55 (average mass) was calculated (Figure 4). This theoretical mass matched precisely the experimentally determined mass of the unmodified protein (see above). Likewise, an average molecular mass of 26,177.69 could be calculated for the gluconoylated form of protein G′e. For the first time, we were able to discover an amino acid sequence elongation that encompassed 46 amino acids at the N-terminus of protein G′e as well as a partial post-translational modification. The deduced amino acid sequence reads into the first 21 amino acids of the protein G sequence (P19909).
Suggested amino acid sequence of protein G′e. Sequence parts covered by MALDI-ISD-MS fragmentation are indicated with arrows below the sequence. Sequence parts covered by ESI-ECD-FT-ICR-MS are indicated with arrows above the sequence. Assigned bond breakages (C″-type and Z′-type ions, respectively) are shown by vertical lines. Boxed amino acid sequence parts were covered by MALDI-QIT-ToF-MS/MS fragmentation analysis. Bold: sequence part of protein G (P19909; aa303–aa497). N-terminal modification by α-N-gluconoylation or α-N-6-phosphogluconoylation, respectively, is indicated by “#”
To test whether the C-terminus of the presumed amino acid sequence was the expected one, we conducted another top-down amino acid sequencing experiment using ESI-ECD-FT-ICR-MS. For that, a nanoESI-MS mass spectrum was recorded under acidic conditions, which showed a series of multiply protonated proteins in the mass range of m/z 700 to 1800. The [M + 26H]26+ ion of m/z 1001.0 (Supplementary Figure 6 and Supplementary Table 11) was isolated and subjected to ECD, giving rise to series of C″ and Z′ ions (Figure 5 and Supplementary Table 12). Detailed ECD fragment analysis showed that from the C-terminus 94 amino acids (aa147–241) could be confirmed (cf. Figure 4). The N-terminal part of the sequence was covered from amino-acid position 1 up to amino-acid position 76. MS/MS error tolerance for ECD fragment ion searching was below 25 ppm. Upon ECD, no cleavage occurred at proline residues, which stands in agreement with literature reports [60].
Top-down sequence analysis of protein G′e by ESI-ECD-FT-ICR MS. (a) Fragment ion spectrum (mass range m/z 250–950) is dominated by C″-type ions (selected ions are labeled). The insert shows the mass range of m/z 550–600. (b) Product ion mass spectrum (mass range m/z 1100–1800) is dominated by Z′-type ions. The insert shows the mass range of m/z 1480–1540. For molecular mass assignment see supplementary Table 12
Protein G′e Sequence Verification
We tested the presumed amino acid sequence of protein G′e (cf. Figure 4) by mass spectrometric peptide mapping. First, protein G′e was digested in solution with trypsin, and peptides were subjected to MALDI-ToF-MS analysis. Almost all the ion signals (i.e., intense and low-abundant ion signals of the resulting mass spectra) were assigned as tryptic peptides of protein G′e with MS error tolerance between 20 and 30 ppm (Figure 6 and Supplementary Table 13; cf. Supplementary Figure 2). Three His-tag containing peptides were observed at m/z 1768.93, 1946.98, and 2026.95, respectively (Supplementary Figure 7 and Supplementary Table 13), confirming partial N-terminal gluconoylation (mass increment of 178.14 Da) and partial α-N-phosphogluconoylation (mass increment of 258.12 Da); consistent with the MALDI-QIT-ToF-MSn sequencing results. Combining partial sequences of all tryptic peptides yielded 100% sequence coverage of protein G′e.
MALDI-ToF-MS analysis of protein G′e peptides derived from tryptic in-solution digestion measured in reflector mode (mass range m/z 950–5000). Insert: linear mode spectrum (mass range m/z 7000–16,000). Selected peptide ion signals are labeled with m/z values. Numbers in parentheses indicate partial sequences of protein G′e. Mass increments of 178 and 258 Da that are due to N-terminal modifications are shown; n.i. denotes not identified. The suggested protein G′e sequence is covered to 100%. For molecular mass assignments see Supplementary Table 13
Peptide mapping of protein G′e by using Asp-N as protease also showed that the suggested amino acid sequence of protein G′e could be matched to the majority of the obtained peptide ion signals (Supplementary Figure 8 and Supplementary Table 14). Again, both, αN-gluconoylation and α-N-6-phosphogluconoylation of the N-terminal peptides were confirmed, and sequence coverage was 100%.
Functional Analysis of Protein G′e
Microscale thermophoresis was performed to define whether the newly determined N-terminal flanking amino acid sequence of protein G′e had an influence on the binding affinity to IgG. For this experiment, the protein G′e concentration was kept constant at ca. 20 nM, whereas the concentration of IgG was varied between 15 pM and 500 nM. From 16 data points, corresponding to different IgG concentrations, the K d value of 9.4 nM was determined for this noncovalent binding in a single experiment (Supplementary Figure 9). Thus, thermophoresis showed that the binding of protein G′e to IgG (from IVIg) was just as strong as that of protein G′ (K d ca. 10 nM) [61]. Accordingly, we conclude that the N-terminal flanking sequence in protein G′e that was added by genetic engineering (and contains a His-tag) does not adversely affect binding properties of protein G′e to IgG.
Discussion
By definition, “de-novo” sequencing (by mass spectrometry) denotes the elucidation of a protein sequence without assistance of a sequence database [62]. A somewhat less stringent version of this definition permits “minimal assistance from genomic data” [63]. One example of a mass spectrometric “de-novo” sequence determination with the help of top-down combined with bottom-up approaches was the determination of the light chain of alemtuzumab, a monoclonal therapeutic antibody [64]. In another example, sequencing of a 21 kDa cytochrome c4 from Thiocapsa roseopersicina was successful by employing a combination of CID and ECD fragmentation experiments on an instrument with a linear ion trap coupled to a Fourier transform-ion cyclotron resonance mass spectrometer [65]. Given that neither genomic data nor precise information about the underlying amino acid sequence from protein G′ were available to us from the starting point and all along during this study, our report presents an actual example of a mass spectrometric “de-novo” sequence elucidation by which the N-terminal flanking amino acid sequence of protein G′e was elucidated.
Only after the complete protein G′e amino acid sequence was experimentally determined were we able to narrow down the likely (commercially available) cloning system that was used for generating the recombinant protein G′e under study. We manually compared the N-terminal flanking amino acid sequence of protein G′e with those amino acid sequences in the lists of pET vectors, which are available for cloning and expressing recombinant proteins in E. coli [66]. The best matching vector-derived amino acid sequence was that of the expression vector pET-28b from Novagen (www.richsinger.com/4402/pET28.pdf). From the ca. 10 possibilities to insert any coding DNA into the multiple cloning site, most likely the Xho I restriction enzyme cleavage site was used. It should be mentioned that without precise knowledge of the amino-acid sequence of the recombinant protein under investigation, finding the correct expression vector was almost impossible because one must pick from over 500 possibilities (i.e., approximately 50 plasmids, each providing multiple cloning sites with typically ca. 10 restriction enzyme recognition sequences).
Given that there are already “minor” structural changes in amino acid sequences—introduced either during genetic engineering or by post-translational modifications—which can cause crucial alterations in the overall functional activity of a protein, precise knowledge of protein primary structures is essential for studies on protein–protein interaction dynamics. For example, a short elongation with just five charged hydrophilic amino acids (KKYPR) at the N-terminus of recombinant human epidermal growth factor caused a significant decrease in its biological activity [67].
Another example of minute structural changes causing significant activity effects is the optimization of pH response and pH sensitivity of the so-called B1 domain of protein G (representing the range of aa47 to aa101 in protein G′e). By targeted mutations, histidine residues were inserted at B1 domain positions 31, 39, and 41, replacing the naturally occurring amino-acid residues glutamine, aspartic acid, and glutamic acid, respectively. This exchange improved binding stability to IgG at higher pH and at the same time caused electrostatic repulsion of protein G from the binding interface of IgG under acidic conditions [68] (residues are highlighted in Supplementary Figure 10). In another protein engineering approach with the so-called C2 domain of protein G (the C2 domain represents aa117 to aa171 in protein G′e), asparagine residues 7 and 36 of the C2 domain [69–71] were substituted with alanine residues to solve a problem of low alkaline stability of protein G. Interaction analysis showed that these amino acid substitutions did not affect the affinity to the Fc fragment of IgG [72]. By contrast, a 50-fold increase in the K d value for IgG-binding (i.e., weakening of the bond) occurs upon a single N34A mutation. Similarly, a K30A mutation results in a 350-fold increase in K d , and a 580-fold increase in the K d occurs with a W42A mutant. Interestingly, the E26A mutant almost abolished binding completely, resulting in approximately a 4000-fold weaker K d value compared with the native B1 domain of protein G [73].
Conclusion
Our study shows that with the help of mass spectrometric “de-novo” sequencing, the primary structure of protein G′e that is available from many companies around the world could be solved completely, revealing a 46-amino acid residue extension at the N-terminus, the presence of an N-terminal His-tag, and a partial gluconoylation. This identification constitutes a first essential step for subsequent studies of protein–protein interactions, which are underway. Although not self-evident, the addition of 46 amino acids at the N-terminus of protein G′e did not cause significant changes in its binding affinity to immunoglobulins.
References
Maxam, A.M., Gilbert, W.: A new method for sequencing DNA. Proc. Natl. Acad. Sci. U. S. A. 74, 560–564 (1977)
Crick, F.H.: Codon–anticodon pairing: the wobble hypothesis. J. Mol. Biol. 19, 548–555 (1966)
Biemann, K.: Laying the groundwork for proteomics: mass spectrometry from 1958 to 1988. J. Proteom. 107, 62–70 (2014)
She, Y.M., Haber, S., Seifers, D.L., Loboda, A., Chernushevich, I., Perreault, H., Ens, W., Standing, K.G.: Determination of the complete amino acid sequence for the coat protein of brome mosaic virus by time-of-flight mass spectrometry. Evidence for mutations associated with change of propagation host. J. Biol. Chem. 276, 20039–20047 (2001)
Catherman, A.D., Skinner, O.S., Kelleher, N.L.: Top down proteomics: facts and perspectives. Biochem. Biophys. Res. Commun. 445, 683–693 (2014)
Hepner, F., Cszasar, E., Roitinger, E., Lubec, G.: Mass spectrometrical analysis of recombinant human growth hormone (Genotropin®) reveals amino acid substitutions in 2% of the expressed protein. Proteome Sci. 3, 1 (2005)
Zhang, H., Ge, Y.: Comprehensive analysis of protein modifications by top-down mass spectrometry. Circ. Cardiovasc. Genet. 4, 711 (2011)
Ezkurdia, I., del Pozo, A., Frankish, A., Rodriguez, J.M., Harrow, J., Ashman, K., Valencia, A., Tress, M.L.: Comparative proteomics reveals a significant bias toward alternative protein isoforms with conserved structure and function. Mol. Biol. Evol. 29, 2265–2283 (2012)
Fermin, D., Allen, B.B., Blackwell, T.W., Menon, R., Adamski, M., Xu, Y., Ulintz, P., Omenn, G.S., States, D.J.: Novel gene and gene model detection using a whole genome open reading frame analysis in proteomics. Genome Biol. 7, R35 (2006)
Teramoto, K., Sato, H., Sun, L., Torimura, M., Tao, H.: A simple intact protein analysis by MALDI-MS for characterization of ribosomal proteins of two genome-sequenced lactic acid bacteria and verification of their amino acid sequences. J. Proteome Res. 6, 3899–3907 (2007)
Kronvall, G.: A surface component in group A, C, and G streptococci with non-immune reactivity for immunoglobulin G. J. Immunol. 111, 1401–1406 (1973)
Guss, B., Eliasson, M., Olsson, A., Uhlén, M., Frej, A.K., Jörnvall, H., Flock, J.I., Lindberg, M.: Structure of the IgG-binding regions of streptococcal protein G. EMBO J. 5, 1567–1575 (1986)
Olsson, A., Eliasson, M., Guss, B., Nilsson, B., Hellman, U., Lindberg, M., Uhlén, M.: Structure and evolution of the repetitive gene encoding streptococcal protein G. Eur. J. Biochem. 168, 319–324 (1987)
Akerström, B., Brodin, T., Reis, K., Björck, L.: Protein G: a powerful tool for binding and detection of monoclonal and polyclonal antibodies. J. Immunol. 135, 2589–2592 (1985)
Alexander, P., Fahnestock, S., Lee, T., Orban, J., Bryan, P.: Thermodynamic analysis of the folding of the streptococcal protein G IgG-binding domains B1 and B2: why small proteins tend to have high denaturation temperatures. Biochemistry 31, 3597–3603 (1992)
Derrick, J.P., Wigley, D.B.: The third IgG-binding domain from streptococcal protein G. An analysis by X-ray crystallography of the structure alone and in a complex with Fab. J. Mol. Biol. 243, 906–918 (1994)
Gallagher, T., Alexander, P., Bryan, P., Gilliland, G.L.: Two crystal structures of the B1 immunoglobulin-binding domain of streptococcal protein G and comparison with NMR. Biochemistry 33, 4721–4729 (1994)
Lian, L.Y., Yang, J.C., Derrick, J.P., Sutcliffe, M.J., Roberts, G.C.K., Murphy, J.P., Goward, C.R., Atkinson, T.: Sequential 1H NMR assignments and secondary structure of an IgG-binding domain from protein G. Biochemistry 30, 5335–5340 (1991)
Sjöbring, U., Björck, L., Kastern, W.: Streptococcal protein G. Gene structure and protein binding properties. J. Biol. Chem. 266, 399–405 (1991)
Akerström, B., Nielsen, E., Björck, L.: Definition of IgG- and albumin-binding regions of streptococcal protein G. J. Biol. Chem. 262, 13388–13391 (1987)
Björck, L., Kastern, W., Lindahl, G., Wideback, K.: Streptococcal protein G, expressed by streptococci or by Escherichia coli, has separate binding sites for human albumin and IgG. Mol. Immunol. 24, 1113–1122 (1987)
Fahnestock, S.R., Alexander, P., Nagle, J., Filpula, D.: Gene for an immunoglobulin-binding protein from a group G streptococcus. J. Bacteriol. 167, 870–880 (1986)
Goward, C.R., Murphy, J.P., Atkinson, T., Barstow, D.A.: xpression and purification of a truncated recombinant streptococcal protein G. Biochem. J. 267, 171–177 (1990)
Ohlson, S., Nilsson, R., Niss, U., Kjellberg, B.M., Freiburghaus, C.: A novel approach to monoclonal antibody separation using high performance liquid affinity chromatography (HPLAC) with SelectiSpher-10 protein G. J. Immunol. Methods 114, 175–180 (1988)
Boström, T., Nilvebrant, J., Hober, S.: Purification systems based on bacterial surface proteins. In: Ahmad, R. (Ed.) Protein Purification, p. 224. InTech, Rijeka, Croatia, (2012)
Hage, D.S.: Affinity chromatography: a review of clinical applications. Clin. Chem. 45, 593–615 (1999)
Kaboord, B., Perr, M.: Isolation of proteins and protein complexes by immunoprecipitation. Methods Mol. Biol. 424, 349–364 (2008)
Nomellini, J.F., Duncan, G., Dorocicz, I.R., Smit, J.: S-layer-mediated display of the immunoglobulin G-binding domain of streptococcal protein G on the surface of Caulobacter crescentus: development of an immunoactive reagent. Appl. Environ. Microbiol. 73, 3245–3253 (2007)
Faulkner, S., Elia, G., Hillard, M., O’Boyle, P., Dunn, M., Morris, D.: Immunodepletion of albumin and immunoglobulin G from bovine plasma. Proteomics 11, 2329–2335 (2011)
Fu, Q., Garnham, C.P., Elliott, S.T., Bovenkamp, D.E., Van Eyk, J.E.: A robust, streamlined, and reproducible method for proteomic analysis of serum by delipidation, albumin and IgG depletion, and two-dimensional gel electrophoresis. Proteomics 5, 2656–2664 (2005)
Björck, L., Blomberg, J.: Streptococcal protein G: a sensitive tool for detection of antibodies to human immunodeficiency virus proteins in Western blot analysis. Eur. J. Clin. Microbiol. 6, 428–429 (1987)
Dancette, O.P., Taboureau, J.L., Tournier, E., Charcosset, C., Blond, P.: Purification of immunoglobulins G by protein A/G affinity membrane chromatography. J. Chromatogr. B Biomed. Sci. Appl. 723, 61–68 (1999)
Zhao, L., Whiteaker, J.R., Pope, M.E., Kuhn, E., Jackson, A., Anderson, N.L., Pearson, T. W., Carr, S.A., Paulovich, A.G.: Quantification of proteins using peptide immunoaffinity enrichment coupled with mass spectrometry. J. Vis. Exp. 53, 1–5 (2011)
Heng, B.C., Aubel, D., Fussenegger, M.: G protein coupled receptors revisited: therapeutic applications inspired by synthetic biology. Annu. Rev. Pharmacol. Toxicol. 54, 227–249 (2014)
Bae, Y.M., Oh, B.K., Lee, W., Lee, W.H., Choi, J.W.: Study on orientation of immunogrlobulin G on protein G layer. Biosens. Bioelectron. 21, 103–110 (2005)
Al-Majdoub, M., Koy, C., Lorenz, P., Thiesen, H.J., Glocker, M.O.: Mass spectrometric and peptide chip characterization of an assembled epitope: analysis of a polyclonal antibody model serum directed against the Sjøgren/systemic lupus erythematosus autoantigen TRIM21. J. Mass Spectrom. 48, 651–659 (2013)
Bradford, M.M.: A rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976)
Kienbaum, M., Koy, C., Montgomery, H.V., Drynda, S., Lorenz, P., Illges, H., Tanaka, K., Kekow, J., Guthke, R., Thiesen, H.J., Glocker, M.O.: MS characterization of apheresis samples from rheumatoid arthritis patients for the improvement of immunoadsorption therapy - a pilot study. Proteom. Clin. Appl. 3, 797–809 (2009)
Al-Majdoub, M., Opuni, K.F., Yefremova, Y., Koy, C., Lorenz, P., El-Kased, R.F., Thiesen, H.J., Glocker, M.O.: A novel strategy for rapid preparation and isolation of intact immune complexes from peptide mixtures. J. Mol. Recogn. 27, 566–574 (2014)
Bantscheff, M., Glocker, M.O.: Probing the tertiary structure of multidomain proteins by limited proteolysis and mass spectrometry. Eur. Mass Spectrom. 4, 279–285 (1998)
Happersberger, H.P., Przybylski, M., Glocker, M.O.: Selective bridging of bis-cysteinyl residues by arsonous acid derivatives as an approach to the characterization of protein tertiary structures and folding pathways by mass spectrometry. Anal. Biochem. 264, 237–250 (1998)
Chen, J.W., Cui, W.D., Giblin, D., Gross, M.L.: New protein footprinting: fast photochemical iodination combined with top-down and bottom-up mass spectrometry. J. Am. Soc. Mass Spectrom. 23, 1306–1318 (2012)
Zubarev, R.A., Kelleher, N.L., McLafferty, F.W.: Electron capture dissociation of multiply charged protein cations. A nonergodic process. J. Am. Chem. Soc. 120, 3265–3266 (1998)
Koy, C., Heitner, J.C., Woisch, R., Kreutzer, M., Serrano-Fernandez, P., Gohlke, R., Reimer, T., Glocker, M.O.: Cryodetector mass spectrometry profiling of plasma samples for HELLP diagnosis: an exploratory study. Proteomics 5, 3079–3087 (2005)
Pecks, U., Seidenspinner, F., Röwer, C., Reimer, T., Rath, W., Glocker, M.O.: Multifactorial analysis of affinity-mass spectrometry data from serum protein samples: a strategy to distinguish patients with preeclampsia from matching control Individuals. J. Am. Soc. Mass Spectrom. 21, 1699–1711 (2010)
El-Kased, R.F., Koy, C., Deierling, T., Lorenz, P., Qian, Z., Li, Y., Thiesen, H.J., Glocker, M.O.: Mass spectrometric and peptide chip epitope mapping of rheumatoid arthritis autoantigen RA33. Eur. J. Mass Spectrom. 15, 747–759 (2009)
El-Kased, R.F., Koy, C., Lorenz, P., Montgomery, H., Tanaka, K., Thiesen, H.J., Glocker, M.O.: A novel Mass spectrometric epitope mapping approach without immobilization of the antibody. J. Proteom. Bioinform. 4, 001–009 (2011)
Happersberger, H.P., Cowgill, C., Glocker, M.O.: Structural characterization of monomeric folding intermediates of recombinant human macrophage-colony stimulating factor beta (rhM-CSF beta) by chemical trapping, chromatographic separation and mass spectrometric peptide mapping. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 782, 393–404 (2002)
Al-Majdoub, M., Opuni, K.F.M., Koy, C., Glocker, M.O.: Facile fabrication and instant application of miniaturized antibody-decorated affinity columns for higher-order structure and functional characterization of TRIM21 epitope peptides. Anal. Chem. 85, 10479–10487 (2013)
Mikkat, S., Koy, C., Ulbrich, M., Ringel, B., Glocker, M.O.: Mass spectrometric protein structure characterization reveals cause of migration differences of haptoglobin a chains in two-dimensional gel electrophoresis. Proteomics 4, 3921–3932 (2004)
Sinz, A., Bantscheff, M., Mikkat, S., Ringel, B., Drynda, S., Kekow, J., Thiesen, H.J., Glocker, M.O.: Mass spectrometric proteome analyses of synovial fluids and plasmas from patients suffering from rheumatoid arthritis and comparison to reactive arthritis or osteoarthritis. Electrophoresis 23, 3445–3456 (2002)
Koy, C., Mikkat, S., Raptakis, E., Sutton, C., Resch, M., Tanaka, K., Glocker, M.O.: Matrix-assisted laser desorption/ionization-quadrupole ion trap-time of flight mass spectrometry sequencing resolves structures of unidentified peptides obtained by in-gel tryptic digestion of haptoglobin derivatives from human plasma proteomes. Proteomics 3, 851–858 (2003)
Röwer, C., Koy, C., Hecker, M., Reimer, T., Gerber, B., Thiesen, H.J., Glocker, M.O.: Mass spectrometric characterization of protein structure details refines the proteome signature for invasive ductal breast carcinoma. J. Am. Soc. Mass Spectrom. 22, 440–456 (2011)
Wienken, C.J., Baaske, P., Rothbauer, U., Braun, D., Duhr, S.: Protein-binding assays in biological liquids using microscale thermophoresis. Nat. Commun. 1, 1–7 (2010)
Jerabek-Willemsen, M., Wienken, C.J., Braun, D., Baaske, P., Duhr, S.: Molecular interaction studies using microscale thermophoresis. Assay Drug Dev. Technol. 9, 342–353 (2011)
Seidel, S.A.I., Dijkman, P.M., Lea, W.A., van den Bogaart, G., Jerabek-Willemsen, M., Lazic, A., Joseph, J.S., Srinivasan, P., Baaske, P., Simeonov, A., Katritch, I., Melo, F.A., Ladbury, J.E., Schreiber, G., Watts, A., Braun, D., Duhr, S.: Microscale thermophoresis quantifies biomolecular interactions under previously challenging conditions. Methods 59, 301–315 (2013)
Roepstorff, P., Fohlman, J.: Proposal for a common nomenclature for sequence ions in mass spectra of peptides. Biomed. Mass Spectrom. 11, 601 (1984)
Hardouin, J.: Protein sequence information by matrix-assisted laser desorption/ionization in-source decay mass spectrometry. Mass Spectrom. Rev. 26, 672–682 (2007)
Geoghegan, K.F., Dixon, H.B., Rosner, P.J., Hoth, L.R., Lanzetti, A.J., Borzilleri, K.A., Marr, E.S., Pezzullo, L.H., Martin, L.B., LeMotte, P.K., McColl, A.S., Kamath, A.V., Stroh, J.G.: Spontaneous alpha-N-6-phosphogluconoylation of a “His tag” in Escherichia coli: the cause of extra mass of 258 or 178 Da in fusion proteins. Anal. Biochem. 267, 169–184 (1999)
Zubarev, R.A.: Electron-capture dissociation tandem mass spectrometry. Curr. Opin. Biotechnol. 15, 12–16 (2004)
Akerström, B., Björck, L.: A physicochemical study of protein G, a molecule with unique immunoglobulin G-binding properties. J. Biol. Chem. 261, 240–247 (1986)
Standing, K.G.: Peptide and protein de novo sequencing by mass spectrometry. Curr. Opin. Struct. Biol. 13, 595–601 (2003)
Seidler, J., Zinn, N., Boehm, M.E., Lehmann, W.D.: De novo sequencing of peptides by MS/MS. Proteomics 10, 634–649 (2010)
Liu, X., Dekker, L.J., Wu, S., Vanduijn, M.M., Luider, T.M., Tolić, N., Kou, Q., Dvorkin, M., Alexandrova, S., Vyatkina, K., Paša-Tolić, L., Pevzner, P.A.: De novo protein sequencing by combining top-down and bottom-up tandem mass spectra. J. Proteome Res. 13, 3241–3248 (2014)
Branca, R.M., Bodó, G., Bagyinka, C., Prokai, L.: De novo sequencing of a 21-kDa cytochrome c4 from Thiocapsa roseopersicina by nanoelectrospray ionization ion-trap and Fourier-transform ion-cyclotron resonance mass spectrometry. J. Mass Spectrom. 42, 1569–1582 (2007)
Rosenberg, A.H., Lade, B.N., Chui, D.S., Lin, S.W., Dunn, J.J., Studier, F.W.: Vectors for selective expression of cloned DNAs by T7 RNA polymerase. Gene 56, 125–135 (1987)
Svoboda, M., Bauhofer, A., Schwind, P., Bade, E., Rasched, I., Przybylski, M.: Structural characterization and biological activity of recombinant human epidermal growth factor proteins with different N-terminal sequences. Biochim. Biophys. Acta 1206, 35–41 (1994)
Watanabe, H., Matsumaru, H., Ooishi, A., Feng, Y.W., Odahara, T., Suto, K., Honda, S.: Optimizing pH response of affinity between protein G and IgG Fc: how electrostatic modulations affect protein-protein interactions. J. Biol. Chem. 284, 12373–12383 (2009)
Frick, I.M., Wikström, M., Forsén, S., Drakenberg, T., Gomi, H., Sjobring, U., Björck, L.: Convergent evolution among immunoglobulin G-binding bacterial proteins. Proc. Natl. Acad. Sci. U. S. A. 89, 8532–8536 (1992)
Gronenborn, A.M., Clore, G.M.: Identification of the contact surface of a streptococcal protein G domain complexed with a human Fc fragment. J. Mol. Biol. 233, 331–335 (1993)
Sauer-Eriksson, A.E., Kleywegt, G.J., Uhlén, M., Jones, T.A.: Crystal structure of the C2 fragment of streptococcal protein G in complex with the Fc domain of human IgG. Structure 3, 265–278 (1995)
Gülich, S., Linhult, M., Ståhl, S., Hober, S.: Engineering streptococcal protein G for increased alkaline stability. Protein Eng. 15, 835–842 (2002)
Sloan, D.J., Hellinga, H.W.: Dissection of the protein G B1 domain binding site for human IgG Fc fragment. Protein Sci. 8, 1643–1648 (1999)
Acknowledgments
The authors express their thanks to Matthias Molnar and Fabian Zehender (NanoTemper Technologies GmbH, Munich, Germany) for providing access to the Monolith NT.115 instrument and for assistance with performing the experiments. They acknowledge the European Union IRSES grant “MS-LIFE” for researcher exchange (PIRSES269256), German Academic Exchange Service (DAAD) for providing a scholarship for YY, and the National Institute of General Medicine of the NIH of the USA (grant no. P41GM103422) for financial support.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 1.42 mb)
Rights and permissions
About this article
Cite this article
Yefremova, Y., Al-Majdoub, M., Opuni, K.F.M. et al. “De-novo” amino acid sequence elucidation of protein G′e by combined “Top-Down” and “Bottom-Up” mass spectrometry. J. Am. Soc. Mass Spectrom. 26, 482–492 (2015). https://doi.org/10.1007/s13361-014-1053-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-014-1053-2