Abstract
A mass spectrometry-based assay combining the specificity of selected reaction monitoring and the protein ion activation capabilities of electron transfer dissociation was developed and employed for the rapid identification of hemoglobin variants from whole blood without previous proteolytic cleavage. The analysis was performed in a robust ion trap mass spectrometer operating at nominal mass accuracy and resolution. Subtle differences in globin sequences, resulting with mass shifts of about one Da, can be unambiguously identified. These results suggest that mass spectrometry analysis of entire proteins using electron transfer dissociation can be employed on clinical samples in a workflow compatible with diagnostic applications.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Hemoglobin is the major protein of red blood cells. It is a tetramer of approximately 64 kDa composed of two α chains and two β (hemoglobin A, or HbA), δ (hemoglobin A2), or γ (hemoglobin F) chains. It plays a major physiological role in carrying oxygen from the lungs to all tissues and organs. Inherited disorders of hemoglobin, also called hemoglobinopathies, are the most common monogenic disorders. It is indeed estimated that 7 % of the world population are mutation carriers. Hemoglobinopathies represent a major health burden with approximately 300,000 children born affected annually [1, 2]. These autosomal recessive genetic diseases are divided into two groups: first, thalassaemias, characterized by the reduction of synthesis of one or more normal globin chains; second, disorders related to the synthesis of a structurally abnormal globin chain (hemoglobin variants). The most common hemoglobin variant is hemoglobin S (HbS) that causes sickle cell disease. Another common variant is hemoglobin C (HbC), responsible for moderate to severe anemia and splenomegaly.
Biological diagnosis of hemoglobinopathies is a complex stepwise procedure involving hematological tests for phenotype determination, liquid chromatography (LC) and electrophoretic methods for the identification and quantification of normal and mutant hemoglobin, and molecular biology techniques to characterize the underlying genetic defect [3]. This multistep process requires multiple analytical laboratory equipments, is often associated with long turnaround times, and must be performed in specialized laboratories by experienced teams. We postulate that top-down mass spectrometry (MS) with electron transfer dissociation (ETD) is a way to simplify and to speed up this complex laboratory procedure. Major improvements were recently brought to MS for protein analysis. Selected reaction monitoring (SRM), an old tool in the MS toolbox, was rediscovered for polypeptide quantification [4]. With SRM, a particular polypeptide ion is first activated into several fragment ions through collision-induced dissociation (CID). The consecutive selection of a particular polypeptide precursor ion (precursor mass) giving rise to a precise fragment ion (fragment mass) within one experiment, referred to as a transition, confers a high specificity to these assays. This methodology can be used for the characterization of hemoglobin variants by mass spectrometry, based on the identification of the polypeptide containing a specific mutation [5]. Although this approach is widely used in research laboratories, most protocols still lack the reproducibility and robustness needed in clinical laboratories [6]. These limitations are largely due to the need of enzymatic digestion prior to MS analysis [7]. Nevertheless, advances in intact protein (top-down) analysis by MS offer a way to circumvent the digestion problem. High-resolution tandem mass spectrometry (MS/MS) of the full-length protein has been previously employed for the characterization of hemoglobin variants [8]. Such methods rely on the accurate and high-resolution detection of multiply charged fragment ions [9, 10]. In parallel, alternative methods to CID were introduced for the analysis of whole proteins. Electron capture dissociation (ECD) was developed in 1998 by Zubarev and co-workers as complementary technique to CID to dissociate peptides and proteins [11]. Due to the rich and specific distribution of peptide backbone fragments obtained with this technique, entire proteins could be sequenced, including identification of post-translational modifications. The more recently introduced ETD uses gas-phase radical anions instead of electrons for the dissociation reaction [12]. Combined with high-resolution instrumentation, ETD was used for the dissociation and structural analysis of proteins up to 150 kDa [13]. While ECD is preferentially performed in a high-end and expensive Fourier transform ion cyclotron resonance mass spectrometers, ETD can be done in less expensive and robust ion trap instruments. Such ion trapping devices can be used with similar modes of operation to SRM, termed SRM-like or pseudo-SRM, to identify and quantify polypeptides over a wide dynamic range [14, 15]. Specific polypeptide precursor ions are selected for activation in the same manner as classical SRM, but the full fragment ion mass spectrum is acquired. Extracted ion chromatograms specific to compounds to be identified and quantified are then reconstructed in silico, once the full dataset is acquired. ETD can be used as an ion activation method in ion trap instruments and hyphenated with this mode of data acquisition [16]. Here, we combined ETD activation and pseudo-SRM techniques for the top-down analysis of full-length proteins, hemoglobin A and hemoglobin variants, from clinical samples. The assay was performed on chromatographic timescale, using a fast ion trap instrument with ETD capabilities and a moderate resolving power.
2 Methods
Whole blood samples were collected in standard EDTA vacutainers at Geneva University Hospitals and stored at 4 °C before processing. All patients gave informed consent. The ethics committee of the Geneva University Hospitals approved this protocol (CE 11-042). A clinical hemoglobin analyzer (Variant II HPLC system with Dual Kit reagents; Bio-Rad, Hercules, CA, USA) was used to perform blood analysis allowing sample characterization in parallel to ETD-MS experiments.
For sample preparation, 250 μL of EDTA blood was centrifuged at 3500 g for 10 min. Then plasma was removed. Red blood cells were suspended in 750 μL of cold solution of 0.9 % NaCl and centrifuged at 700 g for 10 min. The supernatant was removed. This cleaning step was repeated two more times. Then, red blood cells were lysed by the addition of 1750 μL of cold deionized water and centrifuged at 12,000 g for 10 min. At the end of this step, hemoglobin was in solution (supernatant). For direct MS infusion experiments, the sample was desalted with a C18 macro spin column (Harvard Apparatus, Harvard, MA, USA). Briefly, hemoglobin was bound to the spin column with 5 % acetonitrile and 0.1 % formic acid. After washing with the same solution, hemoglobin was eluted with 50 % acetonitrile and 0.1 % formic acid. After evaporation in a vacuum centrifuge, hemoglobin was dissolved in 5 % acetonitrile and 0.1 % formic acid.
Hemoglobin analysis was performed on an Amazon speed ETD mass spectrometer (Bruker Daltonics, Billerica, MA, USA). ETD parameters were optimized to improve globin chains activation. Hemoglobin was directly infused in the mass spectrometer at 1 pmol/μL. Different ETD parameters were tested such as number of reagent anions, ion/ion reaction time, as well as ETD low mass cut-off. The best combination of ETD parameters for hemoglobin were ion accumulation target of 200,000, 400,000 reagent anions (set as ETD ICC), ETD cut-off of at 160 m/z, and reaction time of 60 ms. Three specific fragments ions for hemoglobin A α and β chain, hemoglobin S β chain, and hemoglobin C β chain were selected to allow generating a specific signal for each protein (Table 1). Hemoglobin solutions were injected into a RSLC ultimate 3000 nanoflow ultra high pressure liquid chromatograph (Dionex Thermo Scientific, San Jose, CA, USA) coupled with an amaZon speed ETD ion trap mass spectrometer. The separation was done using a C8 trap column (20 mm, 300 Å particles, inner diameter 100 μm, from Dionex Thermo Scientific) followed by a C8 analytical column (150 mm, 300 Å particles, inner diameter 100 μm, Dionex Thermo Scientific) heated to 34 °C. The proteins were infused in the mass spectrometer through a Captive Spray nanoelectrospray ionization source (Bruker Daltonics). The gradient started with 95 % phase A (water and 0.1 % formic acid) and 5 % of phase B (acetonitrile and 0.1 % of formic acid) and increased to 90 % in 20 min, at a flow rate of 300 nL/min. A volume of 2 μL of hemoglobin solution, at a concentration of 4 pmol/μL was injected. ETD parameters were: ion count control (ICC ETD): 400,000, ion–ion interaction time: 60 ms and cut-off 160 m/z). A scan speed of 32.500 Da/s was used as scan mode, isolation width of 4 m/z and an average of 5 was applied. All experiments were performed in triplicate.
Data were analyzed using Data Analysis software ver. 4.0 (Bruker Daltonics). For LC experiments, extracted ion chromatograms were reconstructed post-acquisition. For High-resolution analysis deconvolution, performed on a qTOF maXis mass spectrometer (Bruker Daltonics), the MaxEntropy algorithm, embedded in the data analysis software, was used.
3 Results and Discussion
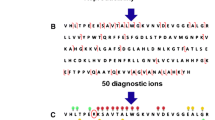
A hemoglobin extract prepared from whole EDTA blood was analyzed using an ion trap mass spectrometer with ETD capabilities. Figure 1a presents a mass spectrum of hemoglobin A, with fully dissociated α and β chains. To perform the most efficient tandem mass spectrometry with ETD, the selected precursor ions should be highly charged because the rate-determining step of the ETD reaction is dependent on the square of the charge of the precursor ion [17]. Also, the m/z distance between adjacent ions should be as wide as possible to allow isolation of one unique chain prior to the dissociation reaction. According to these criteria, the 18 times charged α chain (841.3 m/z) and 19 times charged β chain (836.1 m/z) were chosen for isolation and subsequent ETD. The top-down ETD mass spectrum of the β chain is shown in Figure 1b. With the typical peak width below 0.4 Da, doubly charged fragment ions are unambiguously assigned and fragment ions of the N-terminus are detected up to the position 23 for the wild-type β chain (Figure 1b). Specific fragment ions for α chain, wild-type β chain and mutated β chains of hemoglobin S and C were selected for the next set of experiments (Table 1). In these hemoglobin variants, glutamic acid at position 6 of the β globin chain is replaced by either valine (hemoglobin S) or lysine (hemoglobin C).
ETD-based pseudo-SRM assays were then developed for wild type and mutated hemoglobin chains. For these experiments, hemoglobin was injected into a reversed-phase nanoscale LC column hyphenated to the ion trap mass spectrometer. Three specific fragment ions for each globin chain were used for these assays (Table 1). The absence or presence of a specific fragment ion in the extracted chromatogram unambiguously indicated the presence or absence of a specific chain. An extracted ion chromatogram from a red blood cell extract is shown in Figure 2a. The red blood cells extract was prepared from a patient suffering from sickle cell anemia. The chromatograms clearly show the presence of fragment ions specific to the β chain of hemoglobin S. This patient was transfused with healthy blood before blood sampling and, consequently, hemoglobin A was detected in addition to hemoglobin S. Blood analysis using a clinical hemoglobin analyzer (Variant II HPLC system, Bio-Rad) indicated that hemoglobin S and hemoglobin A concentrations were 70 % and 30 %, respectively. As shown in Figure 2a, this mutated/wild-type β chain ratio was well preserved in chromatograms from the pseudo-SRM assay. The same protocol was then applied on a blood sample from a patient homozygous for hemoglobin C. This single point mutation yields a mass shift of only 1 Da, instead of 30 Da for the hemoglobin S mutation. This very small mass shift prevents discrimination between hemoglobin A and hemoglobin C β chains on the mass spectrum, even with high resolution mass spectrometers (Figure 3). Activation and detection of protein fragment ions was necessary to distinguish the two variants. Accordingly, the pseudo-SRM ETD-MS assay accurately detected the presence of hemoglobin C in patient’s blood (Figure 2b). A chromatographic peak corresponding to three specific transitions of hemoglobin C β chain was indeed clearly visible on the chromatogram (Figure 2b). Because the second isotopic peak of the selected fragments for hemoglobin C β chain overlaps with the fragment ions of non-mutated β chain, a weak specific signal was also detected in the extracted chromatogram corresponding to the β chain of HbA. However, this analytical interference had no impact on the detection of the HbC variant. To demonstrate this, a red blood cell extract from a control patient (HbA) was mixed with a red blood cell extract from a patient homozygote for HbC, simulating a patient heterozygote for HbC. As shown in Figure 2c, HbA and HbC were clearly detected. The isotopic interference corresponding to HbA β chain represented only a small fraction of the ion abundance of HbC β chain (Figure 2b) in the HbC heterozygote patient. The ion abundances are similar for HbA β chain and HbC β chain for this HbC heterozygote scenario.
(a) Reversed-phase nanoHPLC chromatograms of pseudo-SRM ETD assays for hemoglobin α chain, hemoglobin A β chain and hemoglobin S β chain. Blood was collected from a patient homozygous for hemoglobin S transfused with normal blood. The Blood used for this experiment had 30 % of normal hemoglobin (hemoglobin A) and 70 % of mutated hemoglobin (hemoglobin S). In blue, the extracted ion chromatogram of three transitions specific to the α globin chain (841.3 m/z → 600.3 m/z; 728.4 m/z; 829.5 m/z, corresponding to [M + 18 H]18+ → c 1+6 ; c 1+7 ; c 1+8 ion). In red, the extracted ion chromatogram of three transitions specific to the hemoglobin A β chain (836.1 m/z → 694.4 m/z; 823.4 m/z; 951.5 m/z, corresponding to [M + 19 H]19+ → c 1+6 ; c 1+7 ; c 1+8 ion). In green, the extracted ion chromatogram corresponding to three transitions specific to mutated hemoglobin S β chain (transition: 834.5 m/z → 664.4 m/z; 793.5 m/z; 921.5 m/z, corresponding to [M + 19 H]19+ → c 1+6 ; c 1+7 ; c 1+8 ion). (b) Reversed-phase nanoHPLC chromatograms for hemoglobin α chain, hemoglobin A β chain, and hemoglobin C β chain. Blood was collected from a patient homozygous for hemoglobin C. In blue, the extracted ion chromatogram of three transitions specific to the hemoglobin α chain (841.3 m/z → 600.3 m/z; 728.4 m/z; 829.5 m/z, corresponding to [M + 18 H]18+ → c 1+6 ; c 1+7 ; c 1+8 ion). In red, the extracted ion chromatogram of three transitions specific to the hemoglobin A β chain (transition: 836.1 m/z → 694.4 m/z; 823.4 m/z; 951.5 m/z, corresponding to [M + 19 H]19+ → c 1+6 ; c 1+7 ; c 1+8 ion). In orange, the extracted ion chromatogram of three transitions specific to hemoglobin C β chain (836.1 m/z → 693.4 m/z; 822.5 m/z; 950.6 m/z, corresponding to [M + 19 H]19+ → c 1+6 ; c 1+7 ; c8 1+ ion). The small trace in the hemoglobin A β chain corresponds to the second isotopic peak of hemoglobin C fragments. (c) Reversed-phase nanoHPLC chromatograms for hemoglobin α chain, hemoglobin A β chain and hemoglobin C β chain. Red-blood cell extracts from HbA and HbC patients were mixed to simulate a HbC heterozygote. Both chains are now detected with similar ion abundance. The transitions are the same as in (b)
Hemoglobin A and C, extracted from whole blood and analyzed with a high resolution mass spectrometer (qTOF maXis, Bruker Daltonics, Bremen, Germany). (a) Mass spectra of the two hemoglobins with a zoom in the isotopic distribution of hemoglobin β chain at 20+ and 19+ charge states. Upper mass spectrum: hemoglobin A, with the β chain highlighted in light blue. Lower mass spectra: hemoglobin C, with the β chain highlighted in green. The calculated monoisotopic mass (MH+) of HbA β chain is 15858.26 Da, and of HbC β chain is 15857.31 Da. (b) Deconvoluted mass spectra of β chains from the spectra above. The upper spectrum corresponds to the β chain of hemoglobin A, the lower spectrum to the β chain of hemoglobin C. Although high resolution permits the separation of individual isotopes, hemoglobin A and C cannot be distinguished. The observed distortion in the isotopic pattern of the β chain after deconvolution is not explained by the authors.
It is also important to mention that the full tandem mass spectrum is acquired during the pseudo-SRM ETD MS analysis. Thus, specific fragment ions from other sequences and/or hemoglobin mixtures can always be tested post-acquisition to confirm or invalidate the presence of other hemoglobin variants. However, it should be specified that, despite the high specificity of the MS analyzer, the exact identification of some variants can be hampered because different mutations producing different amino acid changes but yielding identical or very similar mass shifts may occur at the same position in a globin chain. For example, hemoglobin Machida, a very rare variant identified in one Japanese family and characterized by the replacement of glutamic acid at position 6 of the β globin chain by a glutamine, yields a mass shift of 0.98. Hemoglobin Machida therefore cannot be distinguished from hemoglobin C (mass shift of 0.95) using the pseudo-SRM ETD MS method with an ion trap. This limitation of MS approaches will have to be considered in future developments for potential clinical use with instruments operating at nominal mass accuracy. However, similar problems are encountered with any screening technique currently used for hemoglobin disorders diagnosis, such ion-exchange chromatography or capillary electrophoresis. Taking in consideration the ethnic origin of the patient and confirmation by molecular biology is the way to address such ambiguous cases.
Finally, the limit of detection of the pseudo-SRM ETD-MS assay was also assessed. Blood from a healthy patient was mixed with blood from a patient homozygous for hemoglobin S in various ratios and analyzed using the same protocol. Results showed that concentrations as low as 4 % of HbS were easily detected (Figure 4).
Reversed-phase nano HPLC chromatograms for hemoglobin β chains. In blue, the extracted ion chromatogram of three transitions specific to hemoglobin A β chain (836.1 m/z → 694.4 m/z; 823.4 m/z; 951.5 m/z, corresponding to [M + 19 H]19+ → c 1+6 ; c 1+7 ; c8 1+ ion). In pink, the extracted ion chromatogram corresponding to three transitions specific to mutated hemoglobin S β chain (transition: 834.5 m/z → 664.4 m/z; 793.5 m/z; 921.5 m/z, corresponding to [M + 19 H]19+ → c 1+6 ; c 1+7 ; c 1+8 ion). (a) Hemoglobin purified from whole blood of a healthy patient was used. As expected, no signal for hemoglobin S β chain could be detected. (b) Whole blood of a healthy patient spiked hemoglobin S to obtain a concentration of 4 % hemoglobin S in 96 % hemoglobin A. The signal for hemoglobin S β chain is clearly visible. The same data processing was applied to both samples
4 Conclusions
Our data show that intact proteins can be specifically analyzed in an ion trap mass spectrometer by pseudo-SRM-ETD. The use of a fast and simple sample preparation protocol combined with a robust, fast, and easy to operate ion trap mass spectrometer allows relatively short turn-around-times (less than 3 hours from blood to results) and makes this methodology potentially suitable for application in clinical laboratories. The power of presented MS-based assay is the possibility to screen for multiple mutations on several polypeptide chains in a single experiment, suggesting that it has the potential for significantly improving diagnostic procedures for hemoglobin variants. Experiments are currently in progress on a cohort of patients with various hemoglobin variants to expand the range of detected mutations. Developments could also be made to allow quantification of normal hemoglobin (A, A2, F) and hemoglobin variants in the same assay.
References
Modell, B., Darlison, M.: Global epidemiology of hemoglobin disorders and derived service indicators. Bull. World Health Org. 86, 480–487 (2008)
Weatherall, D.J.: The inherited diseases of hemoglobin are an emerging global health burden. Blood 115, 4331–4336 (2010)
Old, J.M.: Screening and genetic diagnosis of hemoglobin disorders. Blood Rev. 17, 43–53 (2003)
Lange, V., Picotti, P., Domon, B., Aebersold, R.: Selected reaction monitoring for quantitative proteomics: a tutorial. Mol. Syst. Biol. 4, 1–14 (2008)
Daniel, Y.A., Turner, C., Haynes, R.M., Hunt, B.J., Dalton, R.N.: Rapid and specific detection of clinically significant hemoglobinopathies using electrospray mass spectrometry-mass spectrometry. Br. J. Haematol 130, 635–643 (2005)
Lescuyer, P., Farina, A., Hochstrasser, D.F.: Proteomics in clinical chemistry: will it be long? Trends Biotechnol. 28, 225–229 (2010)
Hoofnagle, A.N.: Quantitative clinical proteomics by liquid chromatography-tandem mass spectrometry: assessing the platform. Clin. Chem. 56, 161–164 (2010)
Light-Wahl, K.J., Loo, J.A., Edmonds, C.G., Smith, R.D., Witkowska, H.E., Shackleton, C.H., Wu, C.S.: Collisionally activated dissociation and tandem mass spectrometry of intact hemoglobin beta-chain variant proteins with electrospray ionization. Biol. Mass Spectrom. 22, 112–120 (1993)
Edwards, R.L., Creese, A.J., Baumert, M., Griffiths, P., Bunch, J., Cooper, H.J.: Hemoglobin variant analysis via direct surface sampling of dried blood spots coupled with high-resolution mass spectrometry. Anal. Chem. 83, 2265–2270 (2011)
Theberge, R., Infusini, G., Tong, W.W., McComb, M.E., Costello, C.E.: Top down analysis of small plasma proteins using an LTQ-Orbitrap. Potential for mass spectrometry-based clinical assays for transthyretin and hemoglobin. Int. J. Mass Spectrom. 300, 130–142 (2011)
Zubarev, R., Horn, D.M., Fridriksson, E.K., Kelleher, N.L., Kruger, N.A., Lewis, M.A., Carpenter, B.K., McLafferty, F.W.: Electron capture dissociation for structural characterization of multiply charged protein cations. Anal. Chem 72, 563–573 (2000)
Syka, J.E.P., Coon, J.J., Schroeder, M.J., Shabanowitz, J., Hunt, D.F.: Peptide and protein sequence analysis by electron transfer dissociation mass spectrometry. Proc. Natl. Acad. Sci. U.S.A. 101, 9528–9533 (2004)
Tsybin, Y.O., Fornelli, L., Stoermer, C., Luebeck, M., Parra, J., Nallet, S., Wurm, F.M., Hartmer, R.: Structural analysis of intact monoclonal antibodies by electron transfer dissociation mass spectrometry. Anal. Chem. 83, 8919–8927 (2011)
Scherl, A., Shaffer, S.A., Taylor, G.K., Kulasekara, H.D., Miller, S.I., Goodlett, D.R.: Genome-specific gas-phase fractionation strategy for improved shotgun proteomic profiling of proteotypic peptides. Anal. Chem. 80, 1182–1191 (2008)
Pak, H., Pasquarello, C., Scherl, A.: Label-free protein quantification on tandem mass spectra acquired in a data-independent mode provides accurate measurements over five orders of concentration magnitude in complex matrices. J. Int. Omics 1, 211–215 (2011)
Wei, B.Y., Juang, Y.M., Lai, C.C.: A novel approach for quantitative peptides analysis by selected electron transfer reaction monitoring. J. Chromatogr. A 1217, 6927–6931 (2010)
Gunawardena, H.P., He, M., Chrisman, P.A., Pitteri, S.J., Hogan, J.M., Hodges, B.D., McLuckey, S.A.: Electron transfer versus proton transfer in gas-phase ion/ion reactions of polyprotonated peptides. J. Am. Chem. Soc. 127, 12627–12639 (2005)
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Coelho Graça, D., Lescuyer, P., Clerici, L. et al. Electron Transfer Dissociation Mass Spectrometry of Hemoglobin on Clinical Samples. J. Am. Soc. Mass Spectrom. 23, 1750–1756 (2012). https://doi.org/10.1007/s13361-012-0446-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-012-0446-3