Abstract
It is now well established that electrospray ionization (ESI) is capable of introducing noncovalent protein assemblies into a desolvated environment, thereby allowing their analysis by mass spectrometry. The degree to which native interactions from the solution phase are preserved in this environment is less clear. Site-directed mutagenesis of FK506-binding protein (FKBP) has been employed to probe specific intra- and inter-molecular interactions within the complex between FKBP and its ligand FK506. Collisional activation of wild-type and mutant-FKBP•FK506 ions, generated by ESI, demonstrated that removal of native protein–ligand interactions formed between residues Asp37, Tyr82, and FK506 significantly destabilized the complex. Mutation of Arg42 to Ala42, or Tyr26 to Phe26 also resulted in lower energy dissociation of the FKBP·FK506 complex. Although these residues do not form direct H-bonds to FK506, they interact with Asp37, ensuring its correct orientation to associate with the ligand. Comparison with solution-based affinity measurements of these mutants has been discussed, including the stabilization afforded by ordered water molecules. Ion mobility spectrometry (IMS) has been employed to provide gas-phase structural information on the unfolding of the complexes. The [M + 6H]6+ complexes of the wild-type and mutants have been shown to resist unfolding and retain compact conformations. However, removal of the basic Arg42 residue was found to induce significant structural weakening of the [M + 7H]7+ complex when raised to dissociation-level energies. Overall, destabilization of the FKBP·FK506 complex, resulting from targeted removal of specific H-bonds, provides evidence for the preservation of these interactions in the desolvated wild-type complex.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Electrospray ionization-mass spectrometry is now widely applied to the analysis of proteins and noncovalent protein complexes. Recent advances have seen its successful application to systems, including the ribosome [1], intact viruses [2, 3], membrane associated protein assemblies [4, 5], and various protein–small molecule ligand complexes [6, 7]. The use of ESI-MS to study noncovalent protein interactions offers several advantages over other biophysical techniques. Notable amongst these are high sensitivity and speed of analysis. A major question concerning the application of MS in studying protein structure and interactions is to what extent the native, solution based, conformation(s) of a protein is preserved following ionization and desolvation by ESI. Significant effort has been directed towards this question [8]. Ion mobility spectrometry (IMS), a gas-phase electrophoretic technique, is now often used in conjunction with MS analyses and allows low resolution structural information of analytes to be directly accessed in the desolvated MS environment [8–14]. These studies have provided insights into gas-phase protein structure, and it is generally accepted that on the timescales of IMS-MS experiments, major structural perturbations can be avoided for low charge state ions [15, 16]. This said, recent studies have shown that low charge states of ubiquitin conformations are significantly different to the crystal structure [17].
At the atomic level, ion mobility studies do not definitively demonstrate that specific noncovalent interactions, the fundamental forces responsible for higher order protein structure in solution, remain intact following ESI and desolvation. One approach to investigate the presence of specific interactions is by systematically removing them and assessing the impact on complex stability. This can be achieved by either introducing single point mutations to the polypeptide chain or by chemically modifying the binding partner/ligand. Klassen and co-workers have utilized both approaches to investigate the interaction between single-chain variable fragment (scFv) of the monoclonal antibody and its native trisaccharide ligand [18–20]. Both experiments provide clear evidence that the specific intermolecular interactions are preserved in these complexes following ESI. Interestingly, these studies were performed using blackbody infrared radiative dissociation (BIRD) as a means of gas-phase dissociation, which allows direct access to time-resolved thermochemical information regarding the protein–ligand binding interaction. These experiments employ activation times of up to seconds, over which significant structural rearrangement may occur [15].
Although BIRD allows quantitative binding information to be obtained in the gas phase, its application has been limited to only a few research groups due to the requirement for FT-ICR instrumentation with a temperature-controlled ICR cell [21]. A more generally available procedure for interrogating protein–ligand stability is to use collision-induced dissociation (CID). Such experiments have been performed in conjunction with genetic/chemical alterations to the protein–ligand binding interface by Douglas and co-workers [22]. In these experiments, collisional activation was achieved in a triple quadrupole instrument and differences in dissociation energy were used to infer relative stability of the gas-phase complexes. Although the results generally complimented the solution data, some discrepancies were also highlighted. For example, in myoglobin, the serine92 residue forms an H-bond interaction with the heme group and subsequent mutation of this residue induces destabilization of the complex in the solution experiments. This was not observed in the gas phase using CID where the results suggest that removing this H-bond leads to stabilization of the interaction. It should be pointed-out that this study used relatively high charge state holo-myoglobin ions ([M + 11H]11+ to [M + 14H]14+), which, by IMS, show multiple and extended conformations (unpublished observation). This may indicate that some of the native interactions in these species were already disrupted prior to collisional activation.
One of the first protein noncovalent interactions to be studied by ESI was that formed between FK-binding protein (FKBP) and its high-affinity ligand, FK506 [23]. FKBP belongs to a superfamily of proteins categorized as peptidyl prolyl cis/trans isomerases (PPIases). FK506 is a 23-membered macrolide (Figure 1b) originating from Streptomyces tsukubaensis [24]. When bound to FKBP, the complex leads to the inhibition of the calcium-dependent phosphatase calcineurin, resulting in immunosuppression [25]. The sub-nanomolar KD FKBP·FK506 complex [26] has been well characterized [27]. Early crystallographic studies by Van Duyne and coworkers identified hydrogen bonds between the ligand and side chain residues Asp37 and Tyr82 [28]. These interactions have been highlighted in Figure 1, along with the additional intramolecular interactions formed by Asp37, which are discussed below. These two interactions have been the subject of solution-based investigations, where the effect of removing them by mutagenesis was examined. A Tyr82Phe mutation resulted in relatively little destabilization of the protein-ligand (P-L) interaction [29]. Free energy measurements showed that this variant is entropically less-favored relative to the wild-type (WT). The more favorable entropic contribution present in the WT-FKBP·FK506 complex is attributed to the displacement of two water molecules upon binding of the ligand. Solution studies have also highlighted the significant contribution of the Asp37-FK506 hydrogen bond in the stability of the P-L complex. An Asp37Val mutant of FKBP resulted in a measured Ki of 350 nM, almost three orders of magnitude lower affinity than that measured for the WT- P-L complex [29].
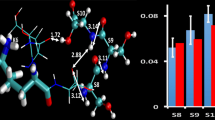
(a) Crystal structure of the FKBP·FK506 complex showing inter-molecular H-bonds between Asp37, Tyr82 (cyan), the FK506 ligand (purple), and intra-molecular H-bonds between Agr42, Tyr26 (pink) and Asp37. From PDB file 1FKJ [46] and rendered in The PyMOL Molecular Graphics System, ver. 0.99, Schrödinger, LLC. (b) The structure of FK506
The Asp37 residue not only forms a direct interaction to the C-10 hydroxyl group of FK506, but also interacts with Tyr26 and Arg42 through H-bonding [28]. This extended network has been studied in solution. A Tyr26Phe mutant resulted in a 37-fold decrease in affinity for FK506 [29]. This is a greater destabilization of the complex than observed for the Tyr82Phe mutant, and is rationalized by the suggested role of Tyr26 in correctly orienting amino acids such as Asp37 [29]. An Arg42Ala-FKBP·FK506 complex exhibited no significant reduction in solution based binding properties relative to the WT, despite the fact that this residue also interacts with Asp37 [28].
Studies presented here were designed to examine whether direct interactions responsible for solution-based binding in the FKBP·FK506 complex are preserved in the gas phase and whether these can be readily observed. Asp37Gly and Tyr82Phe FKBP mutants were produced and the gas-phase stability of their complexes with FK506 were assessed using CID. We have focused on relatively low charge states ([M + 6H]6+ and [M + 7H]7+) to reduce Coulomb-induced unfolding within the protein structure, which has been well documented in previous work [30]. Each mutant resulted in destabilization of the protein–ligand complex. These initial examples were extended to include Arg42 and Tyr26, residues that do not directly interact with FK506, but which do form intramolecular associations with Asp37. Again, these were seen to result in destabilization of the complex. IMS, performed in-line with collisional activation, has been used to offer structural insights. The results are compared qualitatively with solution-based behavior of these mutants.
2 Materials and Methods
2.1 Mass Spectrometry/Ion Mobility
Electrospray ionization-ion mobility-mass spectrometry was performed on a Waters (Altrincham, UK) Synapt High Definition Mass Spectrometer (HDMS), a hybrid quadrupole/ion mobility/orthogonal acceleration time of flight (oa-TOF) instrument. Samples were infused to the standard electrospray (z-spray) source using a Harvard Apparatus Inc. (Edenbridge, Kent, UK) 22 dual syringe pump, model 55-2222 and a 100 μL Hamilton (Bonaduz, GR, Switzerland) syringe connected to a standard Waters ESI probe, at infusion rates between 3 and 5 μL/min. Nanospray was also utilized for some of the acquisitions, using (thin walled) nanospray tips acquired from Waters. The capillary of the ESI source was typically held at voltages between 2.5 and 3 kV, and the nanospray tip at 1–1.8 kV, with the source operating in positive ion mode. The sample cone was operated at 10–20 V. Low voltages were required to avoid gas-phase unfolding and preserve noncovalent interactions. The trap T-wave collisional cell, located just prior to the drift tube, contained argon gas held at a pressure of 2.5 × 10–2 mbar. The ion mobility separator, containing nitrogen gas at 0.45 mbar and ambient temperature, employed a series DC voltage waves (7 V wave height traveling at 280 m/s) to achieve conformational separation. The oa-TOF-MS was operated over the scanning range of m/z 500–3500 at a pressure of 1.8 × 10–6 mbar.
Collisional activation was used to induce protein–ligand dissociation of the FKBP·FK506 complex (WT and mutants). Activation of quadrupole isolated ions was carried out in the trap region (a T-Wave cell located immediately before the ion mobility cell) of the Synapt, by collision with argon gas at 2.5 × 10–2 mbar. The trap collision energy voltage was controlled by the potential offset between the source ion guide and the trap cell, allowing control over translational kinetic energies of ions entering the collision cell. In order to obtain sufficient reproducibility of the data sets, variants were run in parallel, taking at least 5 repeats of 2 min acquisition times at each collision energy. To achieve this, two variants of FKBP were mixed to give approximately equal intensities (counts) and distinguishable m/z values. The MassLynx software was programmed to alternate the quadrupole between isolation of the two species (i.e., one scan at each m/z value). Two variants were run simultaneously and two charge states analyzed each run (6+ and 7+); therefore, the quadrupole was programmed to alternate between four m/z values in each analysis. It was necessary to operate the quadrupole in high resolution settings (LM 14, HM 14), in order to remove adducts from the protonated signal, which lead to destabilizing effects in this complex [31]. This approach accounted for any fluctuations in instrument conditions, by providing internal and external standardization of relative complex stability. Dissociation profiles were constructed by monitoring the percentage precursor remaining (%precursor = precursor counts/(precursor + product counts) × 100) as a function of collision energy. Corrections, based on the small differences in the degree of dissociation between the common complex in each run, were calculated from the off-set in their dissociation profiles. Adjustments were applied so that the data from identical complexes from different runs overlapped, thereby creating a calibrated collision scale across all sets of measurements.
Data were acquired initially at 5 V increments, starting at 3 V. Additional sub-volt increments were acquired close to the 50 % dissociation region, using the quadrupole scanning method described above. Resulting sigmoidal depletion curves were fit using Origin 8 software (Northampton, MA, USA) . Ec50 values, defined as the energy required to induce 50 % dissociation of the complex, were used as quantitative descriptions of dissociation behavior.
Ion mobility measurements were made using the traveling wave mobility cell of the Synapt. Calibration was performed using denatured (49.5:49.5:1 H2O:MeOH:CH3COOH) calibrants bradykinin, ubiquitin, cytochrome c, and myoglobin using a previously reported procedure [32]. Recent studies have demonstrated that using calibrants sprayed from aqueous (nondenaturing) solutions result in more reliable calibration for measurements of ‘native-like’ ions [33]. However, these inaccuracies were shown to become significant when extrapolations to large CCSs are necessary. Calibrant CCSs used in this study ‘bracket’ the values measured for FKBP. Excellent calibration fits were also achieved here and have been included in the Supplementary Information of this manuscript (S1). Collision cross sections were used to estimate the amount of internal energy gained by the ions as they traverse the collision cell, according to the model of Douglas et al. [34]. Briefly, drag coefficients were calculated based on the diffuse scattering model proposed by Henderson [36]. The speed of the ions through the trap collision cell was determined by the speed of the traveling wave in this region (280ms–1 in these studies). The internal energy of the ions were calculated based on the initial injection voltage/energy, the center of mass energy per collision, and the total number of collisions.
2.2 Expression, Purification, and Mutagenesis of FKBP12
The expression vector coding for the WT FKBP12 protein (pGST-FKBP12) was kindly supplied by Dr. Sophie Jackson, University of Cambridge, UK. Recombinant WT FKBP12 protein was expressed and purified as described before [36]. Four mutants, specifically: Y26F, D37G, R42A, and Y82F, were prepared using the WT sequence in the pGST-FKBP12 plasmid as template. Each mutation was introduced by PCR reactions using the QuikChange II site-directed mutagenesis kit (Stratagene, Stockport, Cheshire, UK) with primers containing the appropriate nucleotide substitutions (Sigma, Dorset, UK), the sequences of which are provided in the supplementary information (S2). The presence of the required substitutions and integrity of each sequence after mutagenesis was verified by sequencing before using the plasmids to transform Escherichia coli XL-10 gold cells. Expression and purification of the mutants was performed as for the wild-type FKBP12 protein. Briefly, the GST-tagged protein variants were added to glutathione Sepharose 4B beads (GE Healthcare, Chalfont St. Giles, UK; 100 μL) and thoroughly washed. Following the wash steps, the column was equilibrated in thrombin cleavage buffer (20 mM Tris-HCl, 150 mM NaCl, 2.5 mM calcium chloride dehydrate at pH 8.4) and thrombin was added to the column. Following overnight incubation at room temperature, cleaved protein was eluted from the column. Aliquots (ca. 1 mg ml–1 in thrombin cleavage buffer [see ESI]) were desalted using Vivaspin (Epsom, UK) ultrafilters (MWCO 3000) and diluted to 5 μM in ammonium acetate (25 mM). FK-506 monohydrate was obtained from Sigma (Poole, UK) and was added to the FKBP sample at a 6 μM final concentration.
2.3 Isothermal Titration Calorimetry (ITC)
Isothermal titration calorimetry was performed using a MicroCal Auto ITC instrument. Due to the poor solubility of FK506, both protein and ligand samples were prepared in 10 % propanol. From previous studies with this protein in the presence of other organic modifiers, such as 10 %MeOH [12], the addition of 10 % propanol was not expected to significantly alter the protein structure or the FKBP·FK506 interaction. Each injection mixed 14.03 μL of the ligand solution (200 μM) into the sample cell containing the protein (20 μM) every 3 min. The data were plotted as the molar ratio between the ligand and protein, against the heat change per mole of injected ligand. Fitting was performed iteratively using a standard one-site nonlinear regression model included within Origin 7.0 software.
3 Results
3.1 Direct FKBP-FK506 Interactions
pGST-FKBP variants (WT, Asp37Gly, and Tyr83Phe) were expressed, purified, and cleaved as described in Materials and Methods section. Protein identities were confirmed by ESI-MS analysis (see Supplementary Section S3). The measured mass of WT-FKBP was 12,101.9 Da. Tyr82Phe was measured at 12,086.1 Da (Δm = 15.8; corresponding to removal of an oxygen atom from the WT) and Asp37Gly was measured at 12,044.1 Da (Δm = – 57.8, corresponding to removal of CH2COOH). All proteins contained an additional GS motif on the N-terminus from the thrombin cleavage site. Addition of FK506 (5 μM) to each FKBP variant (5 μM) resulted in a clear complex. Survival of the precursor P–L complex was monitored over a range of collision energies. Ions were accelerated into the ‘Trap’ region of a Waters Synapt mass spectrometer containing Argon collision gas (see section 2). The resulting sigmoidal depletion curves of the Asp37Gly and Tyr82Phe FKBP mutant complexes were compared with those generated for the WT complex. Initial evaluation revealed that the differences in relative stability were very subtle (ΔELAB < 5eV for 6+ and 7+ ions), with the mutants exhibiting lower Ec50 values than the WT FKBP·FK506 complex. We postulated that such minor differences may have simply been due to small variations in instrumental or environmental conditions between each analysis. To remove this uncertainty, acquisitions were performed by measuring protein complexes pair-wise, and switching the quadrupole to transmit each precursor alternately scan-to-scan, as described in the Materials and Methods. Since Asp37Gly possessed the largest mass difference from the WT and Tyr82Phe variants, this was used as a common internal reference in both cases, allowing correction between measurements. Further control of the precision of these experiments was provided by recording multiple measurements at each collision energy to assess the variance in the data.
Measurements were concentrated in the pseudolinear 50 % dissociation regions to allow more precise determination of Ec50 values. The plots of these regions are shown in Figure 2a and b, for the 6+ WT versus Asp37Gly and WT versus Tyr82Phe comparisons respectively. The measured Ec50 values for the 6+ ions of the Asp37Gly, Tyr82Phe, and WT FKBP·FK506 complexes were 193.5, 195.5, and 196.6 eV (Table 1). This demonstrated that the Asp37Gly mutation resulted in greater destabilization of the gas-phase complex compared with Tyr82Phe. The same trend was also observed for the 7+ complexes (see Figure 2c and d, and Table 1). To gain a more quantitative understanding of the degree of destabilization resulting from removal of these interactions, the internal energy (ΔEINT) transferred to the FKBP·FK506 complex ions was estimated using the method of Douglas et al. [34, 37]. The results, expressed as a difference between the WT and mutant Ec50s, termed ΔΔEINT, are shown in Table 1. For both the 6+ and 7+ complexes, larger deviations from the WT dissociation behavior were observed for the Asp37Gly mutant. The ΔΔEINT value for the 6+ charge state of this variant was estimated to be –77 kJ mol–1, whilst that for the Tyr82Phe mutant was –27 kJ mol–1. Hydrogen bond strengths are generally of the order 5–50 kJ mol–1 depending on the nature of the donor and acceptor. Where the acceptor is an anion, as is the case for Asp37, which forms an H-bond to the C10 hydroxyl group of FK506, bond energies tend towards the higher end of the scale. The neutral H-bond between Tyr82 and C8 amide oxygen of FK506 would be expected to be weaker, with a bond energy closer to 10 kJ mol–1. Thus, there is reasonable quantitative agreement between the observed destabilization of the mutant FKBP·FK506 complexes and strength of the H-bonds removed. The 7+ charge states also exhibited similar behavior, although the destabilizing effect of Asp37 removal was somewhat less pronounced at –54 kJ mol–1.
Precursor ion dissociation curves for FKBP·FK506 complexes generated by ESI showing a comparison of wild-type protein and (a) Asp37Gly mutant [M + 6H]6+; (b) Tyr82Phe mutant [M + 6H]6+; (c) Asp37Gly mutant [M + 7H]7+; and (d) Tyr82Phe mutant [M + 7H]7+. Collision energies are ELAB corrected and focused in the region around the Ec50 value. All R2 values > 0.98
In order to examine protein unfolding events during ion activation and dissociation, traveling wave ion mobility spectrometry (TWIMS) measurements were conducted on the 6+ and 7+ charge states of FKBP·FK506. At an ELAB collision energy of 189 eV (chosen because it is close to the Ec50 of all variants/charge states, see Supporting Information Figure S5) the 6+ WT-, Asp37Gly-, and Tyr82Phe-complex ion precursors underwent no significant structural unfolding, and retained a similar compact conformation distribution to the unactivated complex, despite significant levels of P–L dissociation occurring at this energy (see Figure 3a, and Figure S5). This was an interesting result, as it suggested that ligands dissociated from lower charge state ions were ejected from compact structures. Similar results have been reported in protein–protein complexes [38]. In contrast to the 6+ charge state, significantly more structural unfolding was present in the 7+ ions (see Figure 3b). Compared with the unactivated complex, higher drift times were recorded for the ions raised to 189 eV, and multiple conformational distributions were seen. Only relatively minor differences between the drift traces of the activated WT-, Asp37Gly-, and Tyr82Phe-complexes were apparent. These unfolding events may have promoted P–L dissociation, and accounted for the lower Ec50 energies of the 7+ complex, after charge correction (Table 1).
Ion mobility drift trace plots showing activated (189 eV) precursor ions of (a) [M + 6H]6+ and (b) [M + 7H]7+ FKBP·FK506 complex ions for (i) WT-FKBP; (ii) Tyr82Phe-FKBP; (iii) Asp37Gly-FKBP; and (iv) unactivated WT-FKBP as a control. The [M + 6H]6+ charge state shows little or no unfolding, whereas the [M + 7H]7+ complex undergoes significant unfolding
The gas-phase results for Asp37Gly and Tyr82Phe broadly agreed with previous solution-based observations. Relative to WT-FKBP a Asp37Val mutant exhibited a 875-fold decrease in binding affinity for FK506, in solution, which was significantly more than that observed for a Tyr82Phe mutant (2-fold). The Asp37Gly mutant·FK506 complex studied here also exhibited greater destabilization in the gas phase than that of the Tyr82Phe variant. It is notable, however, that the degree of destabilization was significantly different. Entropic effects that drive interactions in solution will be absent from these gas-phase results. The displacement of two water molecules upon formation of the Tyr82–FK506 interaction, for example, is not apparent in the gas phase, and only the enthalpic contribution will be detected. The enthalpic component, itself, will also be altered, due to the major change in dielectric constant upon desolvation.
To establish whether the Asp37Val mutant studied in solution was comparable to the Asp37Gly used here, isothermal titration calorimetry was performed on the Asp37Gly complex (Supplementary Information Figure S6). A KD value of 671 ± 89 nM was determined for the interaction, which is approximately 2-fold higher than the Ki value measured for the Asp37Val variant in previous studies, but of the same order of magnitude. This result confirms the major importance of the Asp37–FK506 interaction in solution.
3.2 Summary – Direct FKBP-FK506 Interactions
It is clear from the data presented that modification of the FKBP Asp37 and Tyr82 side-chains, designed to prevent H-bond interactions with FK506 (at the OH on C-10 and the amide oxygen on C-8 of the ligand, respectively), resulted in destabilization of the P–L complex in the gas-phase. The question arises as to whether this is direct evidence for the preservation of these H-bonds in the desolvated WT-protein complex, or whether mutation may simply compromise protein structural integrity in the gas phase. Asp37 is also involved in intramolecular interactions within the protein structure, and although ion mobility data for the 6+ ion suggest that there is no additional major structural perturbation of this mutant, protein rigidity may be compromised under collisional activation. However, for Tyr82 the situation is much more straightforward. In solution and in the crystal structure, Tyr82 forms an H-bond with the ligand only. Thus, mutation of this residue to Phe would not be expected to destabilize protein structure in any way. Taken together, we believe that these results do provide evidence for the existence of specific interactions between Asp37, Tyr82, and FK506 in the gas phase P–L complex. Comparison with solution-based data reveals that removal of the Asp37 side chain has a much more dramatic effect upon the stability of the complex than does Tyr82 in this environment. In the gas phase, this difference is much less pronounced, but still apparent.
3.3 Intramolecular Protein Interactions
Structural studies have demonstrated that the Tyr26 residue of FKBP does not exhibit an H-bond interaction with FK506. However, ITC measurements have shown that a Tyr26Phe mutation resulted in significant loss in affinity for FK506 (37 fold decrease). This is a much more drastic decrease in affinity than that observed for Tyr82Phe, where a direct interaction with the ligand is disrupted. It has been suggested that Tyr26 is important in ensuring the correct orientation of crucial FK506-interacting residues, such as Asp37. Arg42 is a second residue that interacts with Asp37. The H-bond is of key importance to the correct orientation of the loop consisting of residues 40-44 [39]. Despite this, previous studies designed to remove this polar interaction, via an Arg42Ala mutation, did not appear adversely to affect P–L binding affinity. Indeed, this mutant has 2-fold higher affinity for FK506 than the WT protein, suggesting that Arg42 may actually compete with FK506 for binding to Asp37. In the condensed phase, removal of the Arg42 [39] side chain is compensated for by the presence of two ordered water molecules, which help to stabilize the 40-44 loop. Clearly, this effect would be absent from fully desolvated ions in the gas phase. For these reasons, the two residues, Try26 and Arg42, which both interact with Asp37, were selected as interesting candidates for study. We, therefore, investigated the importance of Tyr26 and Arg42 on the stability of FKBP·FK506 within a desolvated environment under conditions of collisional activation.
As before, following expression and purification of these variants, ESI-MS was used to confirm the presence of mutations and retention of biological activity (ligand binding); see Supplementary Information S4. The 6+ and 7+ variant complexes were isolated and subjected to programmed collisional activation as described earlier. Pairwise measurement of mutants and WT-FKBP was employed to eliminate variation between analyses, and to allow comparison with Asp37Gly and Tyr82Phe mutants, through a common reference. Plots for dissociation of the 6+ Arg42Ala- and Tyr26Phe–FKBP·FK506 mutant complexes are presented in Figure 4a and b, and summarized in Table 1. It is clear that the gas-phase P–L interactions are compromised in these variants. This demonstrates that intramolecular H-bond networks affect the stability of the FKBP·FK506 complex in the gas phase. The Arg42Ala mutant appears more destabilized than the Tyr26Phe, although the differences are small. ΔΔEINT values are –55.0 and –43.4 kJ mol–1, respectively (Table 1). It appears, therefore, that although the Asp37-Arg42 H-bond does not enhance to P–L binding in solution, it has noticeable effect on gas-phase stability.
Precursor ion dissociation curves for FKBP·FK506 complexes generated by ESI showing a comparison of wild-type protein and (a) Arg42Ala mutant [M + 6H]6+; (b) Tyr26Phe mutant [M + 6H]6+; (c) Arg42Ala mutant [M + 7H]7+; and (d) Tyr26Phe mutant [M + 7H]7+. Collision energies are ELAB corrected and focused in the region around the Ec50 value. All R2 values > 0.98
Since residues Arg42 and Tyr26 do not possess any notable interactions with the ligand, the destabilization observed is likely to be caused by weakening of protein tertiary structure. To investigate this, IMS data were collected during the analysis. As for the Asp37Gly and Tyr82Phe mutants, drift traces were compared at 189 eV, since this energy is close to the Ec50 values observed in the experiments and represented the protein conformation immediately before dissociation. For the 6+ charge state all variants possessed near-identical drift times for precursor and at this energy and were indistinguishable from the WT complex under conditions of minimal collisional activation. Therefore, it would seem that little or no permanent unfolding preceded dissociation for the 6+ charge state. This may be useful in structural studies and is analogous to previously reported observations concerning multiprotein assemblies [38]. It is possible that temporary unfolding may occur prior to dissociation but rapid collapse of the structure must ensue, as the dissociated product ion is also compact. It should also be borne in mind that any structural unfolding may also be too subtle to detect using IMS.
Examination of the 7+ FKBP·FK506 complex dissociation data (Figure 4c and Table 1) revealed markedly different behavior from that of the 6+ complex. In particular, significant dissociation of the Arg42Ala-FKBP·FK506 mutant complex was observed. This is the largest destabilization of P–L complex seen in these studies and interestingly originates from removal of an ionic H-bond that does not interact directly with the ligand. This is in stark contrast to solution-based observations, where the Arg42Ala mutation results in no decrease in FK506 affinity [40]. The strength of the ionic H-bond between Asp37 and Agr42 is enhanced in the gas phase and is, therefore, expected to be significant for protein stability in this environment. More importantly, the absence of the two ordered water molecules, seen in the condensed phase structure of this mutant [39], means that there is no compensation for the absence of the guanidine-group of Arg42. This represents a good demonstration that the fundamental interactions responsible for protein structure are altered in the gas phase, especially when discrete ordered water molecules are involved. This also explains why there is a significant difference in stability of the Arg42Ala-FKBP·FK506 complex in solution and in the gas phase. Ion mobility results illustrate that the origin of this destabilization appeared to result from gross structural rearrangement of the complex during activation (Figure 5b). The 7+ precursor ion of the Arg42Ala mutant showed a high degree of unfolding at a kinetic energy (KE) of 189 eV. This behavior was unlike anything seen for the 6+ ion, or any of the other mutants (Figure 5a). Replacing the basic Arg side chain with a nonpolar methyl group (Ala) may have resulted in the additional charge of the 7+ complex being deposited in a location that then compromises the structural stability of the protein in vacuo. An interesting point is that this mutant does not lead to different charge distribution relative to the other variants and suggests that the degree of charging observed in ESI is more dependent on the surface area of the protein in question, in support of previous studies by de la Mora [41] and Kaltashov [42], rather than the amount of basic residues available for protonation. These results highlight that different charge states lead to starkly different gas-phase behavior in protein–ligand complexes, in terms of both stability and structure. It therefore demonstrates that charge state is an important consideration in these experiments. Various new techniques allow greater control over the charge state distributions obtained from ESI [38, 43–45].
Ion mobility drift trace plots showing activated (189 eV) precursor ions of (a) [M + 6H]6+ and (b) [M + 7H]7+ FKBP·FK506 complex ions for (i) WT-FKBP; (ii) Tyr26Phe-FKBP; (iii) Arg42Ala-FKBP; and (iv) unactivated WT-FKBP as a control. The [M + 6H]6+ charge state shows little or no unfolding, whereas the [M + 7H]7+ complex undergoes significant unfolding
3.4 Summary – Intramolecular Protein Interactions
Destabilization of the FKBP·FK506 complex resulting from mutation of Tyr26 and Arg42 has demonstrated that these residues are involved in maintaining integrity of the complex in the gas phase, and provide evidence for a H-bond network with Asp37. IMS results of the 6+ precursor ion complex have shown no obvious unfolding of these mutants, but the 7+ ion of Arg42Ala exhibits significant levels of unfolded protein. This suggests a charge-dependent phenomenon, possibly related to the removal of a basic side chain. Solution studies to establish the affinity of these mutants for FK506 demonstrate that whilst Tyr26 is important, removal of Arg42 has little effect on binding. In fact, it may even weaken the complex. Examination of the crystal structure of WT-FKBP clearly shows that both Try26 and Arg42 interact with Asp37. Removal of the Arg42 guanidine-group can be offset by the presence of two ordered waters, which help to stabilize the 40-44 loop and maintain structural integrity. It is clear from this work that both residues are important in stabilizing the complex in the gas phase.
4 Conclusions
We have assessed the preservation of specific noncovalent interactions formed in a protein–ligand complex in the gas-phase. Collision-induced dissociation has proven to be useful for studying these stability differences. The role of direct H-bonds to the ligand, as well as intramolecular protein interactions have been investigated and found to be significant in stabilizing the FKBP·FK506 complex in the gas phase. This leads us to believe that native H-bonds between Asp37, Tyr82, and FK506, as well as those between Tyr26, Arg42, and Asp37 are probably maintained post-desolvation. It is important to bear in mind that for interactions that depend upon the presence of discrete water molecules, direct comparison is not possible between complexes in solution and those which are fully-desolvated in the gas phase. Taking this effect into account explains the discrepancy between the stability of the Arg42Ala-FKBP·FK506 complex in these two environments. By performing ion mobility measurements in tandem with collisional activation it was possible to monitor any significant structural effects. For the 6+ charge state all mutations showed compact conformations, even at 189 eV, an energy value close to the Ec50. In contrast the 7+ ions exhibited some unfolding at 189 eV and the enhanced unfolding of the 7+ Arg42Ala-FKBP·FK506 complex was readily apparent.
References
Ilag, L.L., Videler, H., McKay, A.R., Sobott, F., Fucini, P., Nierhaus, K.H., Robinson, C.V.: Heptameric (L12)(6)/L10 rather than canonical pentameric complexes are found by tandem MS of intact ribosomes from thermophilic bacteria. Proc. Natl. Acad. Sci. U. S. A. 102, 8192–8197 (2005)
Siuzdak, G., Bothner, B., Yeager, M., Brugidou, C., Fauquet, C.M., Hoey, K., Chang, C.M.: Mass spectrometry and viral analysis. Chem. Biol. 3, 45–48 (1996)
Thomas, J.J., Bothner, B., Traina, J., Benner, W.H., Siuzdak, G.: Electrospray ion mobility spectrometry of intact viruses. Spectros. Int. J. 18, 31–36 (2004)
Wang, S.C., Politis, A., Di Bartolo, N., Bavro, V.N., Tucker, S.J., Booth, P.J., Barrera, N.P., Robinson, C.V.: Ion mobility mass spectrometry of two tetrameric membrane protein complexes reveals compact structures and differences in stability and packing. J. Am. Chem. Soc. 132, 15468–15470 (2010)
Barrera, N.P., Robinson, C.V.: Advances in the mass spectrometry of membrane proteins: From individual proteins to intact complexes. Annu. Rev. Biochem. 80, 247–271 (2011)
Ferguson, P.L., Kuprowski, M.C., Boys, B.L., Wilson, D.J., Pan, J.X., Konermann, L.: Protein folding and protein–ligand interactions monitored by electrospray mass spectrometry. Curr. Anal. Chem. 5, 186–204 (2009)
Sinz, A.: Investigation of protein–ligand interactions by mass spectrometry. Chem. Med. Chem. 2, 425–431 (2007)
Bohrer, B.C., Mererbloom, S.I., Koeniger, S.L., Hilderbrand, A.E., Clemmer, D.E.: Biomolecule analysis by ion mobility spectrometry. Annu. Rev. Anal. Chem. 1, 293–327 (2008)
Clemmer, D.E., Jarrold, M.F.: Ion mobility measurements and their applications to clusters and biomolecules. J. Mass Spectrom. 32, 577–592 (1997)
Hogan Jr., C.J., Ruotolo, B.T., Robinson, C.V., de la Mora, J.F.: Tandem Differential Mobility Analysis-Mass Spectrometry Reveals Partial Gas-Phase Collapse of the GroEL Complex. J. Phys. Chem. B 115, 3614–3621 (2011)
Ruotolo, B.T., Giles, K., Campuzano, I., Sandercock, A.M., Bateman, R.H., Robinson, C.V.: Evidence for macromolecular protein rings in the absence of bulk water. Science 310, 1658–1661 (2005)
Hopper, J.T.S., Oldham, N.J.: Collision induced unfolding of protein ions in the gas phase studied by ion mobility-mass spectrometry: The effect of ligand binding on conformational stability. J. Am. Soc. Mass Spectrom. 20, 1851–1858 (2009)
Jenner, M., Ellis, J., Huang, W.C., Raven, E.L., Roberts, G.C.K., Oldham, N.J.: Detection of a protein conformational equilibrium by electrospray ionization-ion mobility-mass spectrometry. Angew. Chem. Int. Ed. 50, 8291–8294 (2011)
Wyttenbach, T., Bowers, M.T.: Structural stability from solution to the gas phase: Native Solution Structure of Ubiquitin Survives Analysis in a Solvent-Free Ion Mobility-Mass Spectrometry Environment. J. Phys. Chem. B 115, 12266–12275 (2011)
Breuker, K., McLafferty, F.W.: Stepwise evolution of protein native structure with electrospray into the gas phase, 10–12 to 102 s. Proc. Natl. Acad. Sci. U. S. A. 105, 18145–18152 (2008)
Hoaglund, C.S., Valentine, S.J., Clemmer, D.E.: An ion trap interface for ESI-ion mobility experiments. Anal. Chem. 69, 4156–4161 (1997)
Ly, T., Julian, R.R.: Elucidating the tertiary structure of protein ions in vacuo with site-specific photo-initiated radical reactions. J. Am. Chem. Soc. 132, 8602–8609 (2010)
Kitova, E.N., Wang, W.J., Bundle, D.R., Klassen, J.S.: Retention of bioactive ligand conformation in a gaseous protein-trisaccharide complex. J. Am. Chem. Soc. 124, 13980–13981 (2002)
Kitova, E.N., Bundle, D.R., Klassen, J.S.: Evidence for the preservation of specific intermolecular interactions in gaseous protein–oligosaccharide complexes. J. Am. Chem. Soc. 124, 9340–9341 (2002)
Kitova, E.N., Seo, M., Roy, P.N., Klassen, J.S.: Elucidating the intermolecular interactions within a desolvated protein-ligand complex. An experimental and computational study. J. Am. Chem. Soc. 130, 1214–1226 (2008)
Dunbar, R.C.: BIRD (blackbody infrared radiative dissociation): Evolution, principles, and applications. Mass Spectrom. Rev. 23, 127–158 (2004)
Hunter, C.L., Mauk, A.G., Douglas, D.J.: Dissociation of heme from myoglobin and cytochrome b(5): Comparison of behavior in solution and the gas phase. Biochemistry 36, 1018–1025 (1997)
Ganem, B., Li, Y.T., Henion, J.D.: Detection of noncovalent receptor ligand complexes by mass-spectrometry. J. Am. Chem. Soc. 113, 6294–6296 (1991)
Pritchard, D.I.: Sourcing a chemical succession for cyclosporine from parasites and human pathogens. Drug Discov Today 10, 688–691 (2005)
Griffith, J.P., Kim, J.L., Kim, E.E., Sintchak, M.D., Thomson, J.A., Fitzgibbon, M.J., Fleming, M.A., Caron, P.R., Hsiao, K., Navia, M.A.: X-ray structure of calcineurin inhibited by the immunophilin immunosuppressant FKBP12-FK506 complex. Cell 82, 507–522 (1995)
Vanduyne, G.D., Standaert, R.F., Schreiber, S.L., Clardy, J.: Atomic-structure of the rapamycin human immunophilin FKBP-12 complex. J. Am. Chem. Soc. 113, 7433–7434 (1991)
Ivery, M.T.G., Weiler, L.: Modeling the interaction between FK506 and FKBP12: A mechanism for formation of the calcineurin inhibitory complex. Bioorg. Med. Chem. 5, 217–232 (1997)
Vanduyne, G.D., Standaert, R.F., Karplus, P.A., Schreiber, S.L., Clardy, J.: Atomic-structure of fkbp-fk506, an immunophilin-immunosuppressant complex. Science 252, 839–842 (1991)
DeCenzo, M.T., Park, S.T., Jarrett, B.P., Aldape, R.A., Futer, O., Murcko, M.A., Livingston, D.J.: FK506-binding protein mutational analysis: Defining the active-site residue contributions to catalysis and the stability of ligand complexes. Protein Eng. 9, 173–180 (1996)
Clemmer, D.E., Hudgins, R.R., Jarrold, M.F.: Naked protein conformations—cytochrome c in the gas phase. J. Am. Chem. Soc. 117, 10141–10142 (1995)
Hopper, J.T.S., Oldham, N.J.: Alkali metal cation-induced destabilization of gas-phase protein–ligand complexes: Consequences and prevention. Anal. Chem. 83, 7472–7479 (2011)
Ruotolo, B.T., Benesch, J.L.P., Sandercock, A.M., Hyung, S.-J., Robinson, C.V.: Ion mobility-mass spectrometry analysis of large protein complexes. Nat. Protoc. 3, 1139–1152 (2008)
Bush, M.F., Hall, Z., Giles, K., Hoyes, J., Robinson, C.V., Ruotolo, B.T.: Collision cross sections of proteins and their complexes: A calibration framework and database for gas-phase structural biology. Anal. Chem. 82, 9557–9565 (2010)
Chen, Y.L., Campbell, J.M., Collings, B.A., Konermann, L., Douglas, D.J.: Stability of a highly charged noncovalent complex in the gas phase: Holomyoglobin. Rapid Comm. Mass Spectrom. 12, 1003–1010 (1998)
Henderson, C. B.: Drag coefficients of spheres in continuum and rarefield flows. AIAA J. 14, 707–708 (1976)
Main, E.R.G., Fulton, K.F., Jackson, S.E.: Folding pathway of FKBP12 and characterisation of the transition state. J. Mol. Biol. 291, 429–444 (1999)
Mark, K.J., Douglas, D.J.: Coulomb effects in binding of heme in gas-phase ions of myoglobin. Rapid Comm. Mass Spectrom. 20, 111–117 (2006)
Pagel, K., Hyung, S.J., Ruotolo, B.T., Robinson, C.V.: Alternate dissociation pathways identified in charge-reduced protein complex ions. Anal. Chem. 82, 5363–5372 (2010)
Itoh, S., Navia, M.A.: Structure comparison of native and mutant human recombinant FKBP12 complexes with the immunosuppressant drug FK506 (tacrolimus). Protein Sci. 4, 2261–2268 (1995)
Yang, D., Rosen, M.K., Schreiber, S.L.: A composite FKBP12-FK506 surface that contacts calcineurin. J. Am. Chem. Soc. 115, 819–820 (1993)
de la Mora, J.F.: Electrospray ionization of large multiply charged species proceeds via Dole's charged residue mechanism. Anal. Chim. Acta 406, 93–104 (2000)
Kaltashov, I.A., Mohimen, A.: Estimates of protein surface areas in solution by electrospray ionization mass spectrometry. Anal. Chem. 77, 5370–5379 (2005)
Bagal, D., Kitova, E.N., Liu, L., El-Hawiet, A., Schnier, P.D., Klassen, J.S.: Gas phase stabilization of noncovalent protein complexes formed by electrospray ionization. Anal. Chem. 81, 7801–7806 (2009)
Kharlamova, A., Prentice, B.M., Huang, T.Y., McLuckey, S.A.: Electrospray droplet exposure to gaseous acids for the manipulation of protein charge state distributions. Anal. Chem. 82, 7422–7429 (2010)
Hopper, J.T.S., Sokratous, K., Oldham, N.J.: Charge state and adduct reduction in electrospray ionization-mass spectrometry using solvent vapor exposure. Anal. Biochem. 421, 788–790 (2012)
Wilson, K.P., Yamashita, M.M., Sintchak, M.D., Rotstein, S.H., Murcko, M.A., Boger, J., Thomson, J.A., Fitzgibbon, M.J., Black, J.R., Navia, M.A.: Comparative X-ray structures of the major binding-protein for the immunosuppressant FK506 (tacrolimus) in unliganded form and in complex with FK506 and rapamycin. Acta Crystallogr. D 51, 511–521 (1995)
Acknowledgments
The authors thank Katherine Portman and Dr. David Scott for allowing access to ITC equipment. The authors acknowledge funding for this work by the Engineering and Physical Science Research Council (EPSRC) and the School of Chemistry, University of Nottingham.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 7615 kb)
Rights and permissions
About this article
Cite this article
Hopper, J.T.S., Rawlings, A., Afonso, J.P. et al. Evidence for the Preservation of Native Inter- and Intra-Molecular Hydrogen Bonds in the Desolvated FK-Binding Protein·FK506 Complex Produced by Electrospray Ionization. J. Am. Soc. Mass Spectrom. 23, 1757–1767 (2012). https://doi.org/10.1007/s13361-012-0430-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-012-0430-y