Abstract
The fragmentation chemistry of anionic deprotonated hydrogen-deficient radical peptides is investigated. Homolytic photodissociation of carbon–iodine bonds with 266 nm light is used to generate the radical species, which are subsequently subjected to collisional activation to induce further dissociation. The charges do not play a central role in the fragmentation chemistry; hence deprotonated peptides that fragment via radical directed dissociation do so via mechanisms which have been reported previously for protonated peptides. However, charge polarity does influence the overall fragmentation of the peptide. For example, the absence of mobile protons favors radical directed dissociation for singly deprotonated peptides. Similarly, a favorable dissociation mechanism initiated at the N-terminus is more notable for anionic peptides where the N-terminus is not protonated (which inhibits the mechanism). In addition, collisional activation of the anionic peptides containing carbon–iodine bonds leads to homolytic cleavage and generation of the radical species, which is not observed for protonated peptides presumably due to competition from lower energy dissociation channels. Finally, for multiply deprotonated radical peptides, electron detachment becomes a competitive channel both during the initial photoactivation and following subsequent collisional activation of the radical. Possible mechanisms that might account for this novel collision-induced electron detachment are discussed.
Similar content being viewed by others
1 Introduction
Positively charged ions receive the majority of the attention from the mass spectrometric community when it comes to investigating peptides and proteins. Nevertheless, there are important reasons not to ignore anions. For one, many peptides prefer to adopt a negatively charged state. Take for example the synuclein proteins, which are associated with Parkinson’s disease and some forms of cancer, and are dominated by acidic residues [1, 2]. Furthermore, some important post-translational modifications, such as phosphorylation, can also lead to preferential ionization as anionic species [3]. There are also important chemical differences between anions and cations. Most positively charged species are protonated, and dissociation following collisional activation is best explained in terms of the mobile proton model [4]. The absence of mobile protons in anionic peptides is therefore expected to substantially influence the dissociation chemistry. In contrast, most anionic species are deprotonated, with excess electrons serving as the charge carriers. Consequently, detachment of an electron following absorption of a photon occurs much more easily for anions than for cations, which can also potentially influence the resulting dissociation that will be observed [5].
Anionic peptides have been characterized to a limited extent by numerous methods, including collisional activation [6], electron detachment [7], electron capture [8], action spectroscopy [9], and photoelectron spectroscopy [10]. Collisional activation has been the most commonly implemented approach and has been reviewed [11]. Electron detachment can be carried out by both photoactivation and via bombardment with high energy electrons [12]. The overall process leads to conversion of [M – nH]n– to [(M – nH)•](n–1)–, creating a hydrogen-deficient radical peptide (a peptide which contains one less hydrogen atom than its even electron counterpart). Electron capture, or the addition of another electron to a negatively charged peptide, is counterintuitive (but possible) and leads to the creation of a hydrogen abundant radical, which exhibits distinct chemistry [13]. Action spectroscopy in the infrared can be utilized for structural evaluation for peptides of known primary sequence.
Radical directed dissociation (RDD) based methods have enabled unique capabilities for the characterization of peptides and proteins. For example, fragmentation can be directed to particular residues in selected cases [14, 15]. Protein structure can be investigated by monitoring the degree of migration from a specific radical initiation point [16]. The fragmentation pathways and chemistry of positively charged hydrogen-deficient radicals have been studied extensively [17–24]. Hydrogen-deficient radical anions have been neglected by comparison [25, 26]. Interestingly, RDD is not a charge dominated process, in contrast with most other gas-phase chemistry. The primary competition with RDD in positively charged peptides occurs when fully mobile protons exist, creating alternative low energy dissociation channels [27–30]. The absence of mobile protons is therefore potentially significant for RDD in anionic peptides.
It is demonstrated herein that the fragmentation of hydrogen-deficient deprotonated radical peptides is dominated by odd electron chemistry. For singly deprotonated species, RDD fragments including a, z, and c ions, accompanied by various side-chain losses are primarily observed. Competitive b and y ion formation, typical of collisional activation for even electron ions, is not detected. Both of these observations suggest that charges do not play a critical role in defining the fragmentation of radical species. Photoactivation of multiply deprotonated anions generates radical species by both loss of iodine following homolytic cleavage of a carbon–iodine bond, and by electron detachment. Interestingly, collisional activation of the radical following loss of iodine can lead to electron detachment as well. It was also determined that collisional activation of deprotonated peptides with carbon–iodine bonds can be used to site specifically generate radical species, which has not been observed in protonated systems.
2 Experimental
2.1 Chemicals and Reagents
The following peptides were purchased from American Peptide Company (Sunnyvale, CA, USA): ADLIAYL-NH2, DRLYSFGL-NH2, GDGRLYAFGL-NH2, DRVYIHP, YADFVVG-NH2, AEAEYEK, SAEEYEYPS, SQNYPIV, VEPIPY, YPFVEPI, YEVHHQKLVFF, TRSAW, KWDNQ, KNRWEDPGKQLYNVEA, Ac-LMDKEAVYFAHLDIIW, SYSMEHFRWGKPVG, YGGFMRRVGRPEWWMDYQ, YMDGTMSQV. The peptide RGYDARA-NH2 was synthesized by solid phase peptide synthesis. Sodium iodide (NaI), chloramine-T, sodium metabisulfite (Na2S2O5) for peptide iodination, and all the solvents used in the mass spectrometry experiments were purchased from Fisher Scientific (Fairlawn, NJ, USA). Water was purified to 18.2 MΩ resistivity by a Millipore Direct-Q (Millipore, Billerica, MA, USA) prior to use.
2.2 Preparation of Iodinated Peptides
To synthesize iodinated peptides, 10 μL of a 1 mM peptide stock solution (10 nmol) was added to a 1.5 mL Eppendorf test tube containing 90 μL H2O and 10 nmol NaI. To this, 1 μL of 10 mM chloramine-T stock solution (10 nmol) was added and the test tube was allowed to sit in the dark for 1 min. Finally, 15 nmol Na2S2O5 (1.5 μL of the 10 mM stock solution) was quickly spiked into the peptide solution and mixed well to quench the iodination reaction. This resulted in addition of a single iodine to tyrosine in roughly 50% yield. When not quenched, the reaction continues to completion to yield a statistical distribution of singly and multiply iodinated products with a maximum of two iodines added per tyrosine.
2.3 Photodissociation/Collision-Induced Dissociation (PD/CID) of Iodinated Peptides
Aqueous iodinated peptide solutions (0.1 mM) obtained from iodination reactions were first diluted to 10 μM using 50/50 H2O/ACN containing 50 μM ammonium acetate. The dilute iodinated peptide solutions were directly infused into an LTQ linear ion trap mass spectrometer (Thermo Fisher Scientific, Waltham, MA, USA) with a standard ESI source. The electrospray mode was switched between the positive and negative mode to acquire both cationic and anionic peptide ionization and fragmentation. In order to do photodissociation (PD) with the LTQ linear ion trap mass spectrometer, the posterior plate of the LTQ was modified with a quartz window to transmit fourth harmonic (266 nm) laser pulses from a flash lamp pumped Nd/YAG laser (Continuum, Santa Clara, CA, USA). The pulses were synchronized to the end of the isolation step of a typical MS2 experiment by sending a TTL trigger signal from the mass spectrometer to the laser by a digital delay generator (Berkeley Nucleonics, San Rafael, CA, USA). The anionic monoiodinated peptide ions were isolated for PD experiments. Further MS3 experiments were performed by re-isolation of peptide radicals followed by collisional activation. The isolation window width of MS2 and MS3 experiments was limited to 5 Da. The collisional energy was optimized to deplete approximately 75% of the original precursor ion.
2.4 Nomenclature
'M' will be used to describe a molecule or peptide in its natural state. This does not include addition of protons or any chemical modification (i.e. iodination), and assumes an even electron species. Iodinated molecules will be represented with a preceding superscripted label, 'IM'. For protonated peptides, [M + nH]n+ will be used to denote that 'n' protons have been added to the peptide. Similarly, [M – nH]n– indicates a loss of 'n' protons to generate anionic peptides. The creation of radical species by homolytically cleaving a chemical bond via photodissociation is used extensively throughout the manuscript. These radicals are generated independent of charge carrier, location, or polarity and are written in a form to reflect their distonic nature (i.e., [M● + nH]n+ or [M● – nH]n–). Importantly, generation of radicals by this method does not affect the number of protons or charges present in the molecule. Photodetachment is also used to create radicals and does lead to changes in charge state. Photodetachment of an electron from an anionic peptide decreases the charge state, and is accompanied by the formation of a distonic radical. For example, photodetachment of an electron from [M – 2H]2– would produce [M● – 2H]– (note that the number of protons lost and charge state do not match in this case). Finally, both photodissociation and photodetachment can occur for some multiply charged anionic peptides. An example of this would be an iodinated peptide, [IM – 2H]2–, undergoing photodissociation to form [M● – 2H]2– followed by photodetachment to form [M●● – 2H]–. This nomenclature indicates the initial presence of a diradical, although radical-radical recombination may subsequently occur as indicated in the text.
We will adopt the positive ion nomenclature to describe fragments in this manuscript. No structures are implied. The nomenclature is simply used to locate the point of fragmentation and convey whether the charge is on the C-terminus or N-terminus. This is appropriate, as recent careful studies have demonstrated that isobaric fragment ions do not unambiguously have the same structure (i.e., b ions are not all the same) [31, 32].
2.5 Bond Dissociation Energy (BDE) Calculations
DFT calculations were performed at the B3LYP/6-31 G(d) level of theory using Gaussian 09 rev. A.1 (Gaussian, Inc., Wallingford, CT, USA). All BDEs were determined by the isodesmic reaction method where each amino acid was extended on the N-terminus by a carbonyl group and on the C-terminus by an amine. The N-terminus was modeled as an alanine residue extended on the C-terminal side by an amine only. The backbone amide N – H was modeled by the dipeptide Ala-Ala. In the determination of β carbon C-H BDEs, appropriate small molecules with experimentally known BDEs were chosen as references as previously described [37] In addition, (CH3)3CN – H, C6H5 – H, and HCONHCH3 were chosen as reference molecules to accurately model the BDEs of the amine N – H, tyrosyl C – H, and amide N – H radicals respectively [33–35].
3 Results and Discussion
The peptide SAEEYEYPS represents the C-terminal section of procholecystokinin, can be sulfated at both tyrosine residues, and helps regulate satiety [36]. SAEEYEYPS contains three acidic residues, no basic residues, and is easily observed when electrosprayed as a negatively charged ion. Mono-iodination leads to modification at either Tyr5 or Tyr7, roughly in equal abundance. The photodissociation (PD) spectrum generated by exposure to 266 nm light for the singly deprotonated iodinated peptide is shown in Figure 1a. The primary product is the loss of iodine via homolytic cleavage, generating a hydrogen-deficient radical. Some secondary dissociation is also observed, yielding a c8 ion. Importantly, photodetachment of the electron does not appear to be a dominant process in Figure 1a (the ion count does not change significantly after the laser fires). Collisional activation of the peptide radical following loss of iodine and re-isolation is shown in Figure 1b. Abundant side chain dissociation at tyrosine and glutamic acid is observed. The generation of a-type fragments at both tyrosine residues is consistent with a favorable dissociation pathway described previously [37]. Similarly, dissociation on the N-terminal side of serine to produce a c-ion has been described previously. Most of the fragmentation is therefore consistent with previous RDD experiments. However, the z8 ion is not expected based on previously described mechanisms and will be discussed in further detail below. The loss of water is also dominant in Figure 1b, although this loss is not likely to occur via a radical directed mechanism. It has been noted previously that deprotonated peptides containing glutamic acid exhibit water loss [11]. With the exception of water loss, all other notable fragments appear to be generated via radical directed fragmentation mechanisms.
For comparison, the collisional activation of the equivalent protonated radical ion [SAEEYEYPS• + H]+ is shown in Figure 1c. It is clear from the series of b ions which are observed that proton initiated dissociation pathways are competitive with RDD for the protonated peptide. Many of the RDD fragments are similar to those described in Figure 1b, although the z8 fragment is notably absent. Finally, the CID spectra for the deprotonated and protonated even electron species are shown Figure 1d and e, respectively. With the exception of the loss of water, there are no substantial similarities between Figure 1b and d. This suggests that for anionic radical peptides, RDD accounts for the majority of the lowest energy dissociation pathways. In contrast, a sequence of b ions is observed in both Figure 1c and e, confirming that proton initiated dissociation is competitive with RDD when this peptide is protonated. The similarity observed in positive ion mode is consistent with previous results correlating high proton mobility with competitive proton-mediated dissociation [27, 28].
Shown in Figure 2 are CID spectra for deprotonated hydrogen-deficient radicals of YEVHHQKLVFF (an Aβ fragment), ADLIAYL (a neuropeptide), and DRLYSFGL-NH2 (an insect endocrine peptide). There are a variety of side chain losses which are observed (e.g., from Tyr, Leu, Arg, and Glu), which are discussed collectively further below. Backbone fragmentation to yield a and z ions does not occur haphazardly, but is localized primarily to dissociation at aromatic residues, which is consistent with known RDD mechanisms. There are very few b- and y-type ions, which are frequently observed following collisional activation of deprotonated peptides [11]. Taken as a whole, the data in Figures 1 and 2 suggest that RDD dissociation mechanisms for singly deprotonated peptides are largely similar to those described previously in relation to positively charge ions [37]. This observation is perhaps not surprising because no direct role is specified for the charge in any of the previous mechanisms.
Also observed in each spectrum in Figure 2 is a z-type fragment corresponding to loss of the N-terminal residue, as was also noted for SAEEYEYPS in Figure 1. The identities of the N-terminal and n + 1 residues are different for each peptide, suggesting that the dissociation is likely a property of the N-terminus itself. A proposed mechanism to account for these z fragments is shown in Scheme 1. This mechanism begins with the radical located at the N-terminus. This is not unreasonable since the N-terminus has an N–H bond dissociation energy of 383 kJ/mol which is well below that of the initial tyrosyl radical (475 kJ/mol). Migration from the initial tyrosyl radical or from a variety of other potential intermediate sites to the N-terminus would therefore be predicted to be exothermic. Dissociation β to the initial radical eliminates the N-terminal residue and generates a radical x-type fragment. Radical x ions are known to be unstable and frequently spontaneously lose HNCO to yield more stable z-type ions.
This mechanism was probed experimentally with TRSAW, a fragment of the parathyroid hormone related protein [38], as shown in Figure 3. The radical peptides in Figure 3 were generated by noncovalent attachment of an iodonaphthyl 18-crown-6 based molecule as described previously [37]. Collisional activation of [TRSAW● + H]+ yields primarily loss of CO2 and an N-terminal z4 ion, which could be formed by the mechanism shown in Scheme 1. In Figure 3b, TRSAW has been acetylated, yielding a combination of acetylation at the N-terminus and at serine. Collisional activation yields a z4 ion, which retains the acetyl modification, indicating that acetylation of serine does not influence formation of this ion. However, no unmodified z4 ion is detected, suggesting that acetylation at the N-terminus does interfere. According to Scheme 1, N-terminal acetylation would require abstraction of an amide hydrogen, which is significantly less energetically favorable than an amine hydrogen (BDEs of 453 [35] and 383 kJ/mol, respectively). This explains the absence of the z4 ion and also provides a rationale for why z ions are not generated throughout the peptide. In Figure 3b, an unmodified z5 ion is observed. This ion is created by threonine via a mechanism described previously [39] and confirms that the threonine side chain is not acetylated. The mechanism shown in Scheme 1 is again not directly related to charge or charge polarity; however, this pathway is affected if the N-terminus is protonated, which makes hydrogen abstraction significantly less energetically favorable.
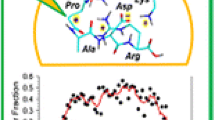
General trends in dissociation observed for deprotonated hydrogen-deficient peptide radicals are summarized in Figure 4 (the peptides are listed in the experimental section). Figure 4a plots the average relative abundance of backbone dissociation for each amino acid. Also shown are the BDEs for the β-hydrogens of each amino acid side chain, which is where backbone dissociation in RDD is typically initiated [17]. As can be seen from the data, lower BDEs (on average) correlate with increased yield of backbone dissociation. This is consistent with observations found previously for positively charged peptides [37]. Importantly, the BDEs only relate to the thermodynamics of radical migration and are, therefore, not expected to correlate quantitatively with the observed dissociation probabilities because kinetics will also play a role in the fragmentation of each peptide. The surprising degree of correlation in Figure 4a suggests that on average for these large molecules, the kinetics of radical migration are not generally dominant.
Statistical summary of data for all peptides listed in the Experimental section. (a) Cβ–H bond dissociation energy (solid square) and normalized relative abundances of anionic radical-induced backbone fragments (solid triangle); on average, backbone fragmentation is favored at residues with low BDEs. (Note: Cys was not present in the peptides sampled.) (b) Propensities of side chain losses at each amino acid following collisional activation of peptide radical anion precursors. Open circles are individual experimentally observed side chain losses, solid triangles represent averaged values. The radical site leading to each loss is indicated by the appropriate Greek letter
In Figure 4b, the relative abundances of side-chain dissociation products are shown, with the average values represented as triangles. Individual values often cover the entire scale and range from no observable dissociation to being the most abundant side chain loss. This range is due to kinetic factors largely dictated by the particular structures for each peptide, which favor or inhibit migration of the radical to a particular side chain. However, the averages are again revealing in terms of the thermodynamics of radical migration. For each side chain from which multiple dissociations are observed, the average loss initiated from the site with a lower BDE [40], which is always the loss originated from a Cα radical (e.g., –43 for Leu) is observed more frequently.
The photoactivation of multiply charged anions is explored in Figure 5. Photoactivation of triply deprotonated iodo-SAEEYEYPS is shown in Figure 5a. Several products are observed, all in fairly low abundance: loss of iodine, loss of an electron, loss of an electron and iodine, and loss of an electron and iodine and CO2. Subsequent collisional activation of the loss of iodine product is shown in Figure 5b. Surprisingly, the primary product afforded by CID is electron detachment [41, 42]. The electron affinities of the initial charge carrying groups (carboxylates) are quite high (>3.2 eV) [43], suggesting that thermionic-like emission is unlikely from these groups. This is supported by the observation that electron detachment is not observed in collisional activation of deprotonated peptide anions. It is also possible that an electron could have been promoted into a weakly bound orbital following absorption of a photon, which is then lost upon collisional activation. However, it is not clear how this electron would survive the high pressure and electric fields present in an ion trap long enough to allow isolation and collisional activation. A third possibility would be radical attack of the carboxylate anion, leading to concurrent formation of a new covalent bond and electron detachment (see Scheme 2). The energy from bond formation could drive electron detachment in this scenario. Collisional activation of the product following electron detachment yields the spectrum shown in Figure 5c, which is consistent with the type of crosslink that would be formed by the mechanism in Scheme 2. Further experiments will be required to definitively determine which of these possibilities actually leads to electron detachment.
Several additional minor side chain losses and backbone fragments consistent with RDD are also observed in Figure 5b. The loss of 71 Da corresponds to loss of the deprotonated glutamic acid side chain as shown in Scheme 3. A small y8 ion is also present. This fragment would not be expected to occur via a radical directed process; however, it is consistent with a cross-linked peptide generated after recombination of a diradical. The y8 ion is the second most abundant product in Figure 5c, suggesting that the y8 in Figure 5b likely results from subsequent fragmentation of hot [SAEEYEYPS●● – 3H]–2 following loss of iodine and electron detachment.
Similar results are obtained for doubly deprotonated YPFVEPI (a casein related peptide) as shown in Figure 5d. Collisional activation of [YPFVEPI● – 2H]–2 (following loss of iodine) again leads primarily to electron detachment. Subsequent activation of the electron detached product yields almost exclusive loss of CO2, which is again consistent with cross-linking following diradical recombination. In addition to electron detachment, loss of CO2 and charged glutamic acid side chains are once again observed.
PD can be utilized to generate radicals regardless of charge polarity; however, collisional activation of iodinated peptides in positive ion mode does not lead to loss of iodine and generation of hydrogen-deficient radical species. Shown in Figure 6a is the CID spectrum for deprotonated IAEAEYEK (iodinated at tyrosine). Surprisingly, the largest peaks correspond to loss of iodine and loss of iodine with several side chains. Isolation of the loss of iodine peak followed by collisional activation is shown in Figure 6b. The corresponding spectrum where the radical was created by photoactivation is provided in Figure 6c. The two figures are very similar, both in terms of product ion identity and intensity, suggesting that the method for creating the radical in this case does not significantly influence the results. The mechanism which leads to loss of iodine in anion mode is not fully understood; however, it is known that the carbon–iodine bond is weak in comparison to naturally occurring bonds in peptides. It is possible that in the absence of proton catalyzed dissociation pathways, cleavage of this bond is simply one of the lowest energy dissociation routes.
4 Conclusions
Many of the radical chemistry dissociation pathways that occur in deprotonated peptides are very similar to those found in protonated peptides. In other words, the charges do not play a central mechanistic role in either case. However, there are also a variety of interesting and important differences that distinguish anionic peptides from their cationic counterparts. For example, the absence of competitive low energy proton initiated dissociation pathways in singly deprotonated radical peptides leads to dissociation dominated by radical chemistry. Similarly, the absence of protons favors backbone fragmentation at the N-terminal residue for anionic radical peptides via a mechanism which is inhibited if the N-terminus is protonated.
In the case of multiply deprotonated peptides, photoactivation at 266 nm yields both fragmentation of carbon–iodine bonds and electron detachment (which is not observed in protonated systems). Interestingly, subsequent collisional activation of the hydrogen-deficient radicals generated by PD also leads to a substantial amount of electron detachment. Such collision-induced electron detachment has not been previously observed for any peptides or peptide radicals to the best of our knowledge. Similarly, collisional activation of deprotonated peptides can be leveraged to create hydrogen-deficient radicals via dissociation pathways yielding homolytic fragmentation of carbon–iodine bonds. Analogous experiments with protonated peptides do not yield radical species.
References
Wright, J.A., Brown, D.R.: ↦-Synuclein and its role in metal binding: Relevance to Parkinson's disease. J. Neurosci. Res. 86(3), 496–503 (2008)
Ahmad, M., Attoub, S., Singh, M.N., Martin, F.L., El-Agnaf, O.M.A.: γ-Synuclein and the progression of cancer. FASEB J. 21(13), 3419–3430 (2007)
Janek, K., Wenschuh, H., Bienert, M., Krause, E.: Phosphopeptide analysis by positive and negative ion matrix-assisted laser desorption/ionization mass spectrometry. Rapid Commun. Mass Spectrom. 15(17), 1593–1599 (2001)
Dongre, A.R., Jones, J.L., Somogyi, A., Wysocki, V.H.: Influence of peptide composition, gas-phase basicity, and chemical modification on fragmentation efficiency: Evidence for the mobile proton model. J. Am. Chem. Soc. 118(35), 8365–8374 (1996)
Larraillet, V., Antoine, R., Dugourd, P., Lemoine, J.: Activated-electron photodetachment dissociation for the structural characterization of protein polyanions. Anal. Chem. 81(20), 8410–8416 (2009)
Hu, P.F., Gross, M.L.: Gas-phase anionic complexes of alkali-metal ions and peptides—structure and collision-activated decompositions. J. Am. Soc. Mass Spectrom. 5(3), 137–143 (1994)
Antoine, R., Joly, L., Tabarin, T., Broyer, M., Dugourd, P., Lemoine, J.: Photo-induced formation of radical anion peptides. Electron photodetachment dissociation experiments. Rapid Commun. Mass Spectrom. 21(2), 265–268 (2007)
Song, H.; Hakansson, K. Improved negative ion electron capture dissociation from coupling with proton transfer reaction. Proceedings of the 59th ASMS Conference on Mass Spectrometry and Allied Topics, 2011.
Oomens, J., Steill, J.D.: The Structure of Deprotonated Tri-Alanine and Its a(3)(–) Fragment Anion by IR Spectroscopy. J. Am. Soc. Mass Spectrom. 21(5), 698–706 (2010)
Vonderach, M., Ehrler, O.T., Matheis, K., Karpuschkin, T., Papalazarou, E., Brunet, C., Antoine, R., Weis, P., Hampe, O., Kappes, M.M., Dugourd, P.: Probing electrostatic interactions and structural changes in highly charged protein polyanions by conformer-selective photoelectron spectroscopy. PCCP 13(34), 15554–15558 (2011)
Bowie, J.H., Brinkworth, C.S., Dua, S.: Collision-induced fragmentations of the (M – H)(–) parent anions of underivatized peptides: An aid to structure determination and some unusual negative ion cleavages. Mass Spectrom. Rev. 21(2), 87–107 (2002)
Kalli, A., Hakansson, K.: Preferential cleavage of S–S and C–S bonds in electron detachment dissociation and infrared multiphoton dissociation of disulfide-linked peptide anions. Int. J. Mass Spectrom. 263(1), 71–81 (2007)
Yoo, H.J., Wang, N., Zhuang, S., Song, H., Hakansson, K.: Negative-ion electron capture dissociation: Radical-driven fragmentation of charge-increased gaseous peptide anions. J. Am. Chem. Soc, Article ASAP (2011). doi:10.1021/ja207736y
Diedrich, J.K., Julian, R.R.: Site-specific radical directed dissociation of peptides at phosphorylated residues. J. Am. Chem. Soc. 130(37), 12212 (2008)
Diedrich, J.K., Julian, R.R.: Site-selective fragmentation of peptides and proteins at quinone-modified cysteine residues investigated by ESI-MS. Anal. Chem. 82(10), 4006–4014 (2010)
Ly, T., Julian, R.R.: Elucidating the Tertiary Structure of Protein Ions in Vacuo with Site Specific Photoinitiated Radical Reactions. J. Am. Chem. Soc. 132(25), 8602–8609 (2010)
Ly, T., Julian, R.R.: Tracking Radical Migration in Large Hydrogen Deficient Peptides with Covalent Labels: Facile Movement does not Equal Indiscriminate Fragmentation. J. Am. Soc. Mass Spectrom. 20(6), 1148–1158 (2009)
Moore, B.N., Blanksby, S.J., Julian, R.R.: Ion-molecule reactions reveal facile radical migration in peptides. Chem. Commun. 33, 5015–5017 (2009)
Hodyss, R., Cox, H.A., Beauchamp, J.L.: Bioconjugates for tunable peptide fragmentation: Free radical initiated peptide sequencing (FRIPS). J. Am. Chem. Soc. 127(36), 12436–12437 (2005)
Chung, T.W., Turecek, F.: Backbone and Side-Chain Specific Dissociations of z Ions from Non-Tryptic Peptides. J. Am. Soc. Mass Spectrom. 21(8), 1279–1295 (2010)
Yin, H., Chacon, A., Porter, N.A., Yin, H.Y., Masterson, D.S.: Free radical-induced site-specific peptide cleavage in the gas phase: Low-energy collision-induced dissociation in ESI- and MALDI mass spectrometry. J. Am. Soc. Mass Spectrom. 18(5), 807–816 (2007)
Wee, S., O'Hair, R.A.J., McFadyen, W.D.: The role of the position of the basic residue in the generation and fragmentation of peptide radical cations. Int. J. Mass Spectrom. 249, 171–183 (2006)
Laskin, J., Futrell, J.H., Chu, I.K.: Is dissociation of peptide radical cations an ergodic process? J. Am. Chem. Soc. 129(31), 9598 (2007)
Chu, I.K., Rodriguez, C.F., Rodriguez, F., Hopkinson, A.C., Siu, K.W.M.: Formation of molecular radical cations of enkephalin derivatives via collision-induced dissociation of electrospray-generated copper (II) complex ions of amines and peptides. J. Am. Soc. Mass. Spectrom. 12(10), 1114–1119 (2001)
Laskin, J., Yang, Z.B., Lam, C., Chu, I.K.: Charge-remote fragmentation of odd-electron peptide ions. Anal. Chem. 79(17), 6607–6614 (2007)
Lam, C.N.W., Chu, I.K.: Formation of anionic peptide radicals in vacuo. J. Am. Soc. Mass Spectrom. 17(9), 1249–1257 (2006)
Laskin, J., Yang, Z.B., Ng, C.M.D., Chu, I.K.: Fragmentation of alpha-Radical Cations of Arginine-Containing Peptides. J. Am. Soc. Mass Spectrom. 21(4), 511–521 (2010)
Knudsen, E.R., Julian, R.R.: Fragmentation chemistry observed in hydrogen deficient radical peptides generated from N-nitrosotryptophan residues. Int. J. Mass Spectrom. 294(2/3), 83–87 (2010)
Lam, A.K.Y., Ryzhov, V., O'Hair, R.A.J.: Mobile Protons Versus Mobile Radicals: Gas-Phase Unimolecular Chemistry of Radical Cations of Cysteine-Containing Peptides. J. Am. Soc. Mass Spectrom. 21(8), 1296–1312 (2010)
Xu, M.J., Song, T., Quan, Q.A., Hao, Q.A., Fang, D.C., Siu, C.K., Chu, I.K.: Effect of the N-terminal basic residue on facile C(↦)–C bond cleavages of aromatic-containing peptide radical cations. Phys. Chem., Chem. Phys. 13(13), 5888–5896 (2011)
Bythell, B.J., Somogyi, A., Paizs, B.: What is the Structure of b(2) Ions Generated from Doubly Protonated Tryptic Peptides? J. Am. Soc. Mass Spectrom. 20(4), 618–624 (2009)
Savitski, M.M., Hith, M., Fung, Y.M.E., Adams, C.M., Zubarev, R.A.: Bifurcating Fragmentation Behavior of Gas-Phase Tryptic Peptide Dications in Collisional Activation. J. Am. Soc. Mass Spectrom. 19(12), 1755–1763 (2008)
Lalevee, J., Allonas, X., Fouassier, J.P.: N–H and ↦ (C–H) bond dissociation enthalpies of aliphatic amines. J. Am. Chem. Soc. 124(32), 9613–9621 (2002)
Blanksby, S.J., Ellison, G.B.: Bond dissociation energies of organic molecules. Acc. Chem. Res. 36(4), 255–263 (2003)
Kaur, D., Kaur, R.P.: Evaluation of N–H bond dissociation energies in some amides using ab initio and density functional methods. J. Mol. Struct. Theochem. 757(1/3), 53–59 (2005)
Vishnuvardhan, D., Beinfeld, M.C.: Role of tyrosine sulfation and serine phosphorylation in the processing of procholecystokinin to amidated cholecystokinin and its secretion in transfected AtT-20 cells. Biochemistry 39(45), 13825–13830 (2000)
Sun, Q.Y., Nelson, H., Ly, T., Stoltz, B.M., Julian, R.R.: Side Chain Chemistry Mediates Backbone Fragmentation in Hydrogen Deficient Peptide Radicals. J. Proteome Res. 8(2), 958–966 (2009)
Philbrick, W.M., Wysolmerski, J.J., Galbraith, S., Holt, E., Orloff, J.J., Yang, K.H., Vasavada, R.C., Weir, E.C., Broadus, A.E., Stewart, A.F.: Defining the roles of parathyroid hormone-related protein in normal physiology. Phys. Rev. 76(1), 127–173 (1996)
Moore, B.N., Ly, T., Julian, R.R.: Radical Conversion and Migration in Electron Capture Dissociation. J. Am. Chem. Soc. 133(18), 6997–7006 (2011)
Rauk, A., Yu, D., Taylor, J., Shustov, G.V., Block, D.A., Armstrong, D.A.: Effects of structure on C- ↦ –H bond enthalpies of amino acid residues: Relevance to H transfers in enzyme mechanisms and in protein oxidation. Biochemistry 38(28), 9089–9096 (1999)
Fu, Y.J., Laskin, J., Wang, L.S.: Collision-induced dissociation of 4Fe–4S cubane cluster complexes: Fe4S4Cl4-x(SC2H5)(x) (2–/1–) (x = 0–4). Int. J. Mass Spectrom. 255, 102–110 (2006)
Simons, J.: Propensity Rules for Vibration-Induced Electron Detachment of Anions. J. Am. Chem. Soc. 103(14), 3971–3976 (1981)
Lu, Z., Continetti, R.E.: Dynamics of the acetyloxyl radical studied by dissociative photodetachment of the acetate anion. J. Phys. Chem. A 108(45), 9962–9969 (2004)
Acknowledgments
The authors gratefully acknowledge funding from the National Science Foundation CHE-0747481.
Author information
Authors and Affiliations
Corresponding author
Additional information
Benjamin Moore and Qingyu Sun contributed equally to this work.
Rights and permissions
About this article
Cite this article
Moore, B., Sun, Q., Hsu, J.C. et al. Dissociation Chemistry of Hydrogen-Deficient Radical Peptide Anions. J. Am. Soc. Mass Spectrom. 23, 460–468 (2012). https://doi.org/10.1007/s13361-011-0318-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-011-0318-2