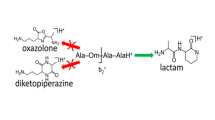
Abstract
Infrared spectra of anionic b-type fragments generated by collision induced dissociation (CID) from deprotonated peptides are reported. Spectra of the b2 fragments of deprotonated AlaAlaAla and AlaTyrAla have been recorded over the 800–1800 cm–1 spectral range by multiple-photon dissociation (MPD) spectroscopy using an FTICR mass spectrometer in combination with the free electron laser FELIX. Structural characterization of the b-type fragments is accomplished by comparison with density functional theory calculated spectra at the B3LYP/6-31++G(d,p) level for different isomeric structures. Although diketopiperazine structures represent the energetically lowest isomers, the IR spectra suggest an oxazolone structure for the b2 fragments of both peptides. Deprotonation is shown to occur on the oxazolone α-carbon, which leads to a conjugated structure in which the negative charge is practically delocalized over the entire oxazolone ring, providing enhanced gas-phase stability.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Collision induced dissociation (CID) tandem mass spectrometry has become an essential tool in proteomics as it is routinely used to determine peptide and protein sequences. In parallel to these application-oriented developments, the reaction chemistry underlying peptide fragmentation has been a topic of much debate over the past decades [1–4]. A good example of where such detailed mechanistic information may aid in the practical analysis of peptide tandem mass spectra are the complications arising from possible sequence scrambling; CID fragments from protonated peptides may adopt cyclic structures in the gas phase, which can reopen at various positions resulting in a permutation of the initial peptide sequence [5–7].
An obviously important parameter in understanding the fragmentation chemistry is knowledge of the molecular structures of the fragment ions formed in the CID process, which can be ambiguous based on the m/z information alone. The combination of tunable infrared lasers with tandem mass spectrometers has made it possible to record IR spectra of CID peptide fragments, which can be related to their structure generally with high confidence. Such detailed structural information in combination with results from computational chemistry can then be used to establish the CID reaction pathways.
Spectroscopic structure determination has thus far been applied predominantly to b-type CID fragment ions generated from protonated peptides. Various different structures have been identified depending on the properties of their amino acid constituents, peptide length, as well as on the collision energy. The b4 fragment ion from protonated leucine-enkephalin was shown to possess an oxazolone structure, which can be protonated at different positions [8, 9]. While b2 fragments had been suggested to form diketopiperazine structures [10], IR spectroscopy definitively determined an oxazolone structure for the b2 fragments from protonated AAA and AGG [11, 12] and a diketopiperazine structure for the b2 ion from HisAla4 [13], For larger b-ions ‘head-to-tail’ cyclization produces macrocyclic structures which may result in sequence scrambling [5, 6, 14, 15]; recent IR spectroscopic studies have indeed revealed the abundant occurrence of such structures [16–18]. Still more recently, a-type fragment ions from protonated peptides were spectroscopically shown to form cyclic structures as well [19, 20]. This behavior of a-type fragments may also lead to sequence scrambling in MSn peptide analysis.
Although most peptide sequencing applications make use of protonated peptide species, current MS instrumentation can generally run in both positive and negative ion mode. Hence, sequence coverage could readily be increased by analyzing the same peptide in negative ion mode and using the complementary information. Moreover, for peptides containing many acidic residues or for phosphorylated peptides, running in negative ion mode, i.e., sequencing the deprotonated peptide, may even be more efficient.
The first IR spectroscopic investigation of deprotonated peptide fragments considered the a3 fragment from Ala3, and the main issue was to determine the deprotonation site in the N-terminal fragment, which no longer possesses a carboxylate moiety at the C-terminus. Resolving the deprotonation site is important as dissociation reactions are generally charge-directed. The IR spectra showed unambiguously that the a –3 fragment has a deprotonated amide (amidate) structure [21] as proposed earlier by Harrison and coworkers [22].
As for protonated peptides, amide bond cleavage is a common peptide fragmentation pattern for deprotonated peptides, leading to the formation of b-type ions. What are the structures of these b-type fragments from deprotonated peptides? Do they also form oxazolone or diketopiperazine structures as is now well-known for protonated peptides? Or are there other structures that are more likely in the case of deprotonated peptides? Moreover, where do these N-terminal fragments deprotonate in the absence of acidic residues? These are the questions that we will address here by comparing experimental IRMPD spectra for deprotonated b2 fragment ions with spectra computed for candidate structures using density functional theory (DFT).
Formation of b-type fragments from deprotonated peptides was first reported by Heerma and co-workers [23, 24], and there are several proposed mechanisms for their formation, which all rely on proton transfer in the first stage of the reaction. Although this “mobile proton theory” may appear counterintuitive for a deprotonated peptide—having one proton fewer, not more—Bache and coworkers showed explicitly that hydrogen atom scrambling also occurs in anionic peptides upon collisional activation [25].
Reaction pathways involving amide bond cleavage as proposed by Bowie and co-workers are depicted in Scheme 1 (for the formation of the b2 ion) and result in a linear amidate b-ion with a C = C = O unit at the C-terminus [26–28]. According to molecular modeling studies at the AM1 level of theory, backbone cleavage leading to b-type ions occurs via an enolate intermediate, which is produced by proton transfer to the COO– moiety [27]. In this mechanism, so called α-cleavage of the peptide backbone leads to an N-deprotonated amino acid (y1 ion), and β-cleavage forms the charged N-terminal fragment (b2 ion).
Proposed mechanism for fragmentation of deprotonated peptides into b and y fragments as suggested by Bowie [27]
An alternative pathway proposed by Harrison and co-workers, depicted in Scheme 2, results in a b2 fragment with a cyclic oxazolone structure [29–32]. Harrison actually proposes two alternative mechanisms. In the first one, the b-ion is formed directly from the parent deprotonated peptide and in the second one, the b2-ion is a sequential fragment formed via the a3 fragment. In Scheme 2, the precursor ion is the deprotonated peptide and an amidate structure is formed by proton transfer to the COO– moiety. This proton transfer is followed by nucleophilic attack of the conjugated negatively charged oxygen atom onto a carbonyl carbon, forming an oxazolone ring, which can retain the negative charge. The same pathway can lead to the y1 fragment if the negative charge remains on the C-terminal fragment. Formation of b2 ions via the a3-fragment also occurs via an amidate intermediate structure [32], see Scheme 3, although this pathway was suggested to be active especially for residues without an amidic hydrogen, such as Pro [29]. The b2 fragment can then be formed via a similar nucleophilic attack.
Mechanism for formation of b2 oxazolone fragments from deprotonated peptides as proposed by Harrison and coworkers [29]
To gain further insight into the possible structures of b2 ions, Harrison and coworkers turned to computational chemistry [33]. In this study, the energetically most favorable b2 ion structures (for the GG sequence) correspond to our structures b2AA_1, b2AA_8, b2AA_9, b2AA_11, and b2AA_12 presented in Figure 1. Inspired by Harrison’s findings, we here report on the results of an IR spectroscopic investigation of the b2 ion derived from the deprotonated peptides tri-alanine (AAA) and alanyl-tyrosil-alanine (AYA).
2 Experimental and Computational Methods
All experiments were conducted using the free electron laser FELIX [34] and a home-built FTICR mass spectrometer equipped with a 4.7 T actively-shielded superconducting magnet (Cryomagnetics Inc., Oak Ridge, TN, USA) [35–37].
Deprotonated peptide anions were generated by electrospray ionization (Micromass Z-Spray source) using solutions containing about 1 mM of the peptide and 1 mM NaOH to enhance the formation of peptide anions. Tri-alanine and alanyl-tyrosyl-alanine were obtained from Sigma-Aldrich (Zwijndrecht, The Netherlands) and Bachem (Weil am Rhein, Germany), respectively. A mixture of methanol and water (7:3 ratio) was used as solvent. CID fragments were generated by nozzle-skimmer dissociation in the high-pressure region at the inlet of the mass spectrometer. Ions were accumulated in a linear hexapole trap held at a background pressure of about 10–3 Torr, where the trapping voltages can be adjusted to further enhance the formation of a specific CID ion. The ions were then transferred to the ICR cell, where the anion of interest (b2) was isolated using a SWIFT excitation pulse [38]. Mass-isolated ions were irradiated with the tunable light from the free electron laser [34]. FELIX produces 5 μs long pulses with an energy of ~60 mJ (~30–40 mJ in the ICR cell) and a bandwidth of ~0.5% of the central wavelength. In the current experiments, the FELIX wavelength was scanned from 5.5 to 13 μm, which corresponds to a spectral range of 800–1800 cm–1. Ions were irradiated for 3.8 s, while the entire FTICR sequence lasted 6 s. Four mass spectra were averaged at each wavelength point. Infrared multiple-photon dissociation (IR-MPD) occurs whenever the frequency of the laser is in resonance with a particular vibration in the ion [39–42]; tens to hundreds of photons are absorbed leading to the dissociation of the anion. Recording the photofragment yield as a function of wavelength gives an IR spectrum, where the yield is determined as \( \sum {{\text{I}}\left( {fragments} \right)} /\sum {{\text{I}}\left( {all{\,\, }ions} \right)} \).
Experimental spectra were compared with calculated spectra to identify the molecular structures. Optimized anion structures and computed spectra were generated by DFT using the B3LYP functional and the 6-31++G(d,p) basis set, implemented in Gaussian 03. This method was previously found to give a good prediction for the energies and IR spectra for deprotonated species [43, 44]. Starting structures for the b2 ions were constructed on the basis of those previously investigated by Harrison and coworkers [33] and using chemical intuition. Structures exhibiting resonance stabilization by conjugation and with a high number of H-bonding interactions were considered to be the most attractive candidates. The structures are presented in Figure 1. Computed harmonic frequencies are scaled by 0.97 and convoluted with a 25 cm–1 Gaussian line shape function to facilitate easy comparison with experimental spectra.
3 Results
The precursor peptides were selected on the basis of their activity and residue size. Trialanine was chosen because the alanine residue is nonpolar and relatively chemically inert to possible involvement of the side chain in the fragmentation reaction. Alanyl-tyrosyl-alanine possesses a bulky tyrosine residue which might influence the reaction by steric interactions. More importantly, the tyrosine hydroxyl group can deprotonate relatively easily (similar to phenol, pKaTyr ~10) and may thus compete for the negative charge with other acidic protons in the system.
3.1 Alanyl-Alanyl-Alanine
The b2 fragment anion of deprotonated AAA at m/z 141 was generated by in-source CID, in particular by manipulating the settings of the cone voltage and the hexapole accumulation trap, and subsequently mass-isolated in the ICR cell. Upon IR activation the b2 anion was found to undergo dissociation forming fragments with m/z 111, 113, 42, and 97 (ordered by their abundance), suggesting loss of formaldehyde (H2CO), CO, methyl substituted oxazolone and CO2, respectively. Observation of the CO2 loss channel suggests that two oxygen atoms have become bonded to the same carbon atom in the b2-fragment ion. This fragmentation pathway was also observed, though with CID of N-benzoylpeptides, in the work of Harrison and coworkers [30] and was interpreted as an indication for an oxazolone structure. These data provide circumstantial evidence for an oxazolone structure of the b2 fragment.
Figure 2 presents a direct comparison of the experimental spectrum with those computed for the various candidate structures of Figure 1. It is immediately obvious that only one of the suggested structures shows good agreement with the experimental spectrum, which is structure b2AA_1, corresponding to a 2,4-substituted-5-oxazolone. The calculations formally place the negative charge for this structure on the α-carbon atom that has become integrated in the oxazolone ring. The dominant band observed in the IRMPD spectrum at 1670 cm–1 corresponds to the oxazolone C = O stretching mode. Since the structure is in fact an ester, one could have expected the carbonyl stretching mode to occur at slightly higher wave numbers. However, the enol resonance induces a substantial red-shifting of this band. Bands with lower intensities appear at 1532, 1331, 1233, and 1096 cm–1 and correspond to N = C stretching, ring vibration, C-N in-ring stretching, and NH2 twisting, respectively.
Experimental IRMPD spectrum of the b2 fragment of deprotonated AAA (black line) compared with calculated spectra (red and blue dashed line) for various oxazolone, diketopiperazine, and linear candidate structures (in order of increasing relative energy). The red arrow indicates the deprotonation site in each of the structures
Calculated relative energies are given for each of the suggested structures in the upper left corner of each panel in Figure 2. Even though the spectrum of structure b2AA_1 clearly fits best with the experimental spectrum, there are two structures with substantially lower energies. These are the diketopiperazine structures deprotonated on the ring nitrogen and carbon atoms, which are lower in energy than b2AA_1 by 41 and 6 kJ/mol, respectively. It is obvious in the experimental spectrum that only one C = O stretching mode is present, while for the diketopiperazine structures two C = O stretching modes are predicted.
To further support our structural assignment and to exclude the presence of diketopiperazine structures, the spectrum of commercially available cyclic-dialanine, which has a diketopiperazine structure, was recorded. The results are shown in Figure 3.
Comparison of the experimental IRMPD spectra of deprotonated cyclo-dialanine (red), b2 from AAA (black dashed line), and b2 from AYA (blue dash-dotted line). The red-dotted line represents the calculated spectrum for the lowest-energy diketopiperazine structure, which shows that cyclo-Ala2 is deprotonated on one of the nitrogen atoms
The experimental spectrum of deprotonated cyclic-dialanine shows good agreement with the spectrum calculated for the N-deprotonated diketopiperazine structure (see Figure S1 in the Supporting Information). The experimental spectrum indeed shows two carbonyl stretching modes, which occur at different frequencies because of the different molecular environment of the two carbonyl groups in the diketopiperazine ring. The band at 1660 cm–1 is due to the carbonyl stretch which is not affected by the amidate resonance, whereas the feature at 1590 cm–1 is due to the carbonyl involved in the amidate resonance, which gives partial single bond character to this CO bond.
It is further noticed that IRMPD dissociation channels are different from those observed for the b2 fragment. Main fragment mass channels for cyclic-dialanine are m/z 42 and 97. While the m/z 42 fragment may correspond to NCO– or NH = CH-CH –2 , an exact mass determination suggests the former stoichiometry. Mass 97 is due to the loss of a neutral mass 44 fragment, which could either be acetaldehyde (CH3CHO) or carbon dioxide (CO2); here the exact mass clearly suggests acetaldehyde loss, which is in line with experimental data by Bowie and coworkers on the dissociation of deprotonated 2,5-diketopiperazines indicating that CO2-loss is unlikely [45]. In conclusion, both the spectrum and the fragmentation channels resulting from IRMPD are different for the AlaAla b2 anion and the deprotonated cyclo-dialanine, providing independent experimental evidence against a diketopiperazine structure for the b2 fragment.
3.2 Alanyl-Tyrosyl-Alanine
The b2 fragment generated by front-end CID from deprotonated AYA is observed at m/z 233. Upon IR activation the b2 anion was found to undergo dissociation into fragments at m/z 97, 93, 42, and 88 (ordered by their abundance). Mass 97 may correspond to the loss of neutral 4-methylen-2,5-cyclohexadiene-1-one followed by the loss of formaldehyde, while mass 93 likely corresponds to the formation of the phenoxide (C6H5O–) anion. Both of these fragmentation channels are commonly observed for anionic peptides containing a tyrosine residue [46]. The identity of the m/z 88 fragment is not clear. Although its mass corresponds to that of deprotonated alanine, this would be unusual since the b2 ion from AYA does not possess a carboxylate moiety, and moreover does not possess a C-terminal alanine residue. The analysis of the IRMPD spectrum of the b2 fragment described below is independent of the identity of the fragment ions observed.
Our main motivation for recording the IRMPD spectrum of the b2 from AYA was to confirm the robustness of the deprotonated oxazolone motif found for the b2 fragment from deprotonated AAA. Specifically, deprotonation may in this case occur on the tyrosine side chain forming a phenoxide anion. Hence, we can qualitatively compare the gas-phase acidity of the oxazolone motif with that of phenol. In addition, we recorded the IRMPD spectrum for the intact deprotonated AYA anion to establish the initial deprotonation site of the parent anion. Note that the deprotonation site of the Tyr amino acid has been the subject of substantial discussion recently [47–49]. Figure S3 in the Supporting Information unambiguously establishes a carboxylate structure for deprotonated AYA, analogous to what we found previously for deprotonated Tyr using IR spectroscopy [49].
The IRMPD spectrum recorded for the AYA b2 fragment anion is shown in Figure 4, compared with calculated spectra for two oxazolone isomers, with deprotonation occurring either on the oxazolone α-carbon or on the phenol moiety of the Tyr side-chain. One can immediately see from the comparison that this b2 fragment possesses a deprotonated oxazolone structure, analogous to the b2 fragment of deprotonated AAA. Comparisons with spectra calculated for other possible structures can be found in the Supporting Information (Figure S2). The most intense absorption observed at 1675 cm–1 is attributed to the oxazolone C = O stretching mode, which appears slightly red-shifted at 1655 cm-–1 in the calculated spectrum. Figure 3 shows that the experimentally observed frequency is consistent between the b2 ions from AYA and AAA with a difference of only 5 cm–1, suggesting that the discrepancy with theory may be an artifact of the harmonic frequency calculation. Assignment of the structure as a deprotonated oxazolone rather than a phenoxide anion is furthermore consistent with calculated relative free energies given for each of the structure in Figure 4.
Apart from the oxazolone carbonyl stretching mode, there are more similarities between the spectra of the b2 ions from AYA and AAA. Absorption features observed at 1539, 1327 and 1231 cm–1 in the spectrum of b2 from AYA can be correlated to absorptions at 1532, 1331, 1233 cm–1 in the spectrum of b2 from AAA (see Figure 3). According to the calculations, these peaks are due to the C = N stretching vibration, the oxazolone ring vibration and the oxazolone C-N stretching mode, respectively. In addition, features observed at 833, 809, 1080, and 1168 cm–1 in the experimental spectrum of b2 from AYA correspond to bands at 854, 824, 1080, and 1161 cm–1, respectively, in the theoretical spectrum and are due to aromatic out-of-plane and in-plane C-H vibrations. Furthermore, a band of special interest located at 1153 cm–1 in the calculations is observed as a shoulder at 1156 cm–1 in the experiment. This peak is assigned as the hydroxyl COH bending mode of the phenol moiety and is very close in frequency to the analogous mode in the IRMPD spectrum of deprotonated tyrosine [49]. This similarity provides further experimental evidence for the tyrosine side-chain not being deprotonated. In addition, the feature at 1223 cm–1 (1218 cm–1 in the theoretical spectrum) is due to the phenolic C–O stretching mode. Finally, we note that the NH2 bending vibration calculated at 912 cm–1 is not well reproduced in the experimental spectrum. Previous IRMPD spectra of deprotonated amino acids have shown that the frequency and intensity of this mode are often found to deviate strongly from their theoretical values [49], which we suspect to be due to the very anharmonic potential for this vibration [50, 51]. All experimental and theoretical data thus strongly suggest that both deprotonated peptides form b2 fragments with an oxazolone structure and that deprotonation occurs on the α-carbon embedded in the oxazolone ring.
4 Discussion
One may now wonder what makes this particular oxazolone structure more attractive than all other candidate structures presented in Figure 1. As described above, the oxazolone ring is believed to be formed by nucleophilic attack of the N-terminal amide oxygen onto the neighboring amide carbon (see Scheme 2) [29], whereas the thermodynamically more favored diketopiperazine structure would have to be formed by nucleophilic attack of the N-terminal nitrogen onto the second amide carbon. This latter reaction requires a trans to cis isomerization of the N-terminal amide bond before cleavage of the peptide bond occurs. For protonated peptides, rotation around the amide bond faces a relatively low energetic penalty due to the reactive configuration in which the mobile proton has migrated to the amide nitrogen, reducing the bond order of the amide bond. However, while energetically feasible, this isomerization is entropically unfavorable [52–56], preventing the formation of diketopiperazine structures. Although no transition state calculations were performed for deprotonated peptides, in addition to possible entropic constraints, trans-to-cis isomerization likely faces higher energetic barriers, since in the reactive configuration where the amide moiety is deprotonated (Scheme 2), the double-bond character of the amide bond is enhanced rather than reduced. For oxazolone structures to be formed on the other hand, only rotation around the N–Cα bond is needed. Furthermore, formation of the diketopiperazine structure may require deprotonation of the N-terminal amino group for it to become sufficiently nucleophilic, which is however energetically unfavorable in the gas phase as it would strongly localize the negative charge. Trans-to-cis isomerization would then have to occur around a neutral, conjugated amide bond facing a substantial thermodynamic barrier. These considerations make formation of the diketopiperazine an unlikely reaction pathway and provide qualitative arguments for fragmentation pathways leading to oxazolone structures.
Of all possible oxazolone structures in Figure 1, the structure identified in the experiments has the lowest energy and here we rationalize why this structure is particularly favorable. A close inspection of this structure in Figure 5 reveals that there is a high level of charge delocalization in this oxazolone b2 structure, in which the negative charge can be delocalized over five heavy atoms. None of the other oxazolone forms in Figure 1 allows one to draw more than just two reasonable resonance structures. This conjecture can be further supported with calculated bond lengths of the b2AA_1 deprotonated structure compared with those for its neutral counterpart (Figure 6). Elongation of the Ca = O bond by 0.046 Å and shortening of the Ca–Cc bond by 0.126 Å is a consequence of the enolate resonance (red isomer in Figure 5). On the other hand, substantial contraction of the Cc–N bond occurs (0.075 Å) as well as an elongation of the N = Cb bond (0.029 Å) as a result of the imine resonance (shown as the blue isomer in Figure 5). This results in a conjugated structure for the b2AA_1 anion as presented in violet in Figure 5.
Finally, we note that the fragment ion structures that have been identified in this and a previous study [21] on the fragmentation of deprotonated tripeptides support the conclusions of Harrison, Siu, and coworkers [29, 32, 33]. In particular, we showed that the a3 fragment of deprotonated AAA has an amidate structure that corresponds to the suggested structure and the fragmentation pathway of Harrison shown here in Scheme 3. Our results for the b2 fragment anions described in the current study are in support of Harrison’s suggested b-ion structure, possibly formed via the a3 anion (see Scheme 3).
5 Conclusions
The first spectroscopic investigation of anionic b-ions has been presented; gas-phase IRMPD spectra for the b2 fragments of the deprotonated peptides AAA and AYA have been recorded. Comparisons with calculated spectra show unambiguously that the b2 fragments generated from both peptides have an oxazolone structure, which is deprotonated on the α-carbon in the oxazolone ring. No evidence was found for b2 fragments having the lower-energy diketopiperazine structure, which was further confirmed by recording an experimental spectrum for deprotonated cyclo-dialanine that is substantially different from that recorded for the CID generated b2 ion of deprotonated AAA. The deprotonated oxazolone b-ion structure identified here is shown to be a robust motif in the sense that it is also formed in the presence of other acidic protons, in this case on the tyrosine side chain.
Formation of the deprotonated oxazolone structure is likely due to an energetically and/or entropically favorable transition state as compared with that for the formation of the diketopiperazine structure. The latter pathway requires a trans-to-cis isomerization of the peptide bond, which possibly represents a higher energetic barrier compared with an analogous isomerization in protonated peptides, although computational investigations are required to substantiate this qualitative argument. Furthermore, of all possible oxazolone structures that can be formed, the one where deprotonation occurs on the oxazolone Cα is particularly stable due to a high degree of charge delocalization, which is an important factor in the gas-phase stability of ions.
In more general terms, this study shows that characterization of CID peptide fragment structures by IRMPD spectroscopy is also successful for anionic species. A deeper understanding of peptide fragmentation in negative ion mode may eventually lead to an increased application in peptide sequencing strategies.
References
McLafferty, F. W. Tandem Mass Spectrometry (MS/MS): A Promising New Analytical Technique for Specific Component Determination in Complex Mixtures. Acc. Chem. Res. 13, 33–39 (1980)
Siuzdak, G.: The Emergence of Mass-Spectrometry in Biochemical-Research. Proc. Nat. Acad. Sci. U.S.A. 91, 11290–11297 (1994)
Yates, J.R.: Mass Spectrometry and the Age of the Proteome. J. Mass Spectrom. 33, 1–19 (1998)
Biemann, K.: Contributions of Mass Spectrometry to Peptide and Protein Structure. Biol. Mass Spectrom. 16, 99–111 (1988)
Harrison, A. G. Peptide Sequence Scrambling through Cyclization of b5 Ions. J. Am. Soc. Mass Spectrom. 19, 1776–1780 (2008)
Bleiholder, C., Osburn, S., Williams, T.D., Suhai, S., Van Stipdonk, M., Harrison, A.G., Paizs, B.: Sequence-Scrambling Fragmentation Pathways of Protonated Peptides. J. Am. Chem. Soc. 130, 17774–17789 (2008)
Yu, L., Tan, Y., Tsai, Y., Goodlett, D.R., Polfer, N.C.: On the Relevance of Peptide Sequence Permutations in Shotgun Proteomics Studies. J. Proteome Res. 10, 2409–2416 (2011)
Polfer, N.C., Oomens, J., Suhai, S., Paizs, B.: Spectroscopic and Theoretical Evidence for Oxazolone Ring Formation in Collision-Induced Dissociation of Peptides. J. Am. Chem. Soc. 127, 17154–17155 (2005)
Polfer, N.C., Oomens, J., Suhai, S., Paizs, B.: Infrared Spectroscopy and Theoretical Studies on Gas-Phase Protonated Leu-Enkephalin and Its Fragments: Direct Experimental Evidence for the Mobile Proton. J. Am. Chem. Soc. 129, 5887–5897 (2007)
Savitski, M.M., Falth, M., Fung, Y.M.E., Adams, C.M., Zubarev, R.A.: Bifurcating Fragmentation Behavior of Gas-Phase Tryptic Peptide Dications in Collisional Activation. J. Am. Soc. Mass Spectrom. 19, 1755–1763 (2008)
Oomens, J., Young, S., Molesworth, S., van Stipdonk, M.: Spectroscopic Evidence for an Oxazolone Structure of the b2 Fragment Ion from Protonated Tri-Alanine. J. Am. Soc. Mass Spectrom. 20, 334–339 (2009)
Yoon, S.H., Chamot-Rooke, J., Perkins, B.R., Hilderbrand, A.E., Poutsma, J.C., Wysocki, V.H.: IRMPD Spectroscopy Shows that AGG Forms an Oxazolone b +2 Ion. J. Am. Chem. Soc. 130, 17644–17645 (2008)
Perkins, B.R., Chamot-Rooke, J., Yoon, S.H., Gucinski, A.C., Somogyi, A.R.D., Wysocki, V.H.: Evidence of Diketopiperazine and Oxazolone Structures for HA b +2 Ion. J. Am. Chem. Soc. 131, 17528–17529 (2009)
Molesworth, S., Osburn, S., Van Stipdonk, M.: Influence of Size on Apparent Scrambling of Sequence During CID of b-Type Ions. J. Am. Soc. Mass Spectrom. 20, 2174–2181 (2009)
Molesworth, S., Osburn, S., Van Stipdonk, M.: Influence of Amino Acid Side Chains on Apparent Selective Opening of Cyclic b5 Ions. J. Am. Soc. Mass Spectrom. 21, 1028–1036 (2010)
Chen, X., Yu, L., Steill, J.D., Oomens, J., Polfer, N.C.: Effect of Peptide Fragment Size on the Propensity of Cyclization in Collision-Induced Dissociation: Oligoglycine b2–b8. J. Am. Chem. Soc. 131, 18272–18282 (2009)
Chen, X. A.; Steill, J. D.; Oomens, J.; Polfer, N. C. Oxazolone Versus Macrocycle Structures for Leu-Enkephalin b2–b4: Insights from Infrared Multiple-Photon Dissociation Spectroscopy and Gas-Phase Hydrogen/Deuterium Exchange. J. Am. Soc. Mass Spectrom. 21, 1313–1321 (2010)
Erlekam, U., Bythell, B.J., Scuderi, D., Van Stipdonk, M., Paizs, B., Maitre, P.J.: Infrared spectroscopy of fragments of protonated peptides: direct evidence for macrocyclic structures of b5 ions. Am. Chem. Soc. 131, 11503–11508 (2009)
Verkerk, U.H., Siu, C.-K., Steill, J.D., El Aribi, H., Zhao, J., Rodriquez, C.F., Oomens, J., Hopkinson, A.C., Siu, K.W.M.: a2 Ion Derived from Triglycine: An N1-Protonated 4-Imidazolidinone. J. Phys. Chem. Lett. 1, 868–872 (2010)
Bythell, B.J., Maitre, P., Paizs, B.: Cyclization and Rearrangement Reactions of an Fragment Ions of Protonated Peptides. J. Am. Chem. Soc. 132, 14766–14779 (2010)
Oomens, J., Steill, J.D.: The Structure of Deprotonated Tri-Alanine and Its a –3 Fragment Anion by IR Spectroscopy. J. Am. Soc. Mass Spectrom. 21, 698–706 (2010)
Chass, G.A., Marai, C.N.J., Harrison, A.G., Csizmadia, I.G.: Fragmentation Reactions of a2 Ions Derived from Deprotonated Dipeptides—a Synergy between Experiment and Theory. J. Phys. Chem. A 106, 9695–9704 (2002)
Kulik, W., Heerma, W.: Fast Atom Bombardment Tandem Mass-Spectrometry for Amino-Acid Sequence Determination in Tripeptides. Biomed. Environm. Mass Spectrom. 18, 910–917 (1989)
Vansetten, D., Kulik, W., Heerma, W.: Isomeric Tripeptides—a Study on Structure Spectrum Relationship. Biomed. Environ. Mass Spectrom. 19, 475–480 (1990)
Bache, N., Rand, K.D., Roepstorff, P., Ploug, M., Jørgensen, T.J.D.: Hydrogen Atom Scrambling in Selectively Labeled Anionic Peptides Upon Collisional Activation by MALDI Tandem Time-of-Flight Mass Spectrometry. J. Am. Soc. Mass Spectrom. 19, 1719–1725 (2008)
Waugh, R.J., Bowie, J.H.: A Review of the Collision-Induced Dissociations of Deprotonated Dipeptides and Tripeptides—an Aid to Structure Determination. Rapid Commun. Mass Spectrom. 8, 169–173 (1994)
Bowie, J.H., Brinkworth, C.S., Dua, S.: Collision-Induced Fragmentations of the (M – H)– Parent Anions of Underivatized Peptides: An Aid to Structure Determination and Some Unusual Negative Ion Cleavages. Mass Spectrom. Rev. 21, 87–107 (2002)
Eckersley, M., Bowie, J.H., Hayes, R.N.: Collision-Induced Dissociations of Deprotonated Peptides, Dipeptides and Tripeptides with Hydrogen and Alkyl α-Groups—an Aid to Structure Determination. Org. Mass Spectrom. 24, 597–602 (1989)
Harrison, A. G.; Siu, K. W. M.; El Aribi, H. Amide Bond Cleavage in Deprotonated Tripeptides: A Newly Discovered Pathway to ''b2 Ions. Rapid Commun. Mass Spectrom. 17, 869–875 (2003)
Harrison, A., Young, A.: Fragmentation of Deprotonated N-Benzoylpeptides: Formation of Deprotonated Oxazolones. J. Am. Soc. Mass Spectrom. 15, 446–456 (2004)
Harrison, A.G.: Sequence-Specific Fragmentation of Deprotonated Peptides Containing H or Alkyl Side Chains. J. Am. Soc. Mass Spectrom. 12, 1–13 (2001)
Harrison, A.G., Young, A.B.: Fragmentation Reactions of Deprotonated Peptides Containing Proline. The Proline Effect. J. Mass Spectrom. 40, 1173–1186 (2005)
Chass, G.A., Marai, C.N.J., Setiadi, D.H., Csizmadia, I.G., Harrison, A.G.: A Hartree-Fock, MP2 and DFT Computational Study of the Structures and Energies of ''b2 Ions Derived from Deprotonated Peptides. A Comparison of Method and Basis Set Used on Relative Product Stabilities. J. Mol. Struct. Theochem. 675, 149–162 (2004)
Oepts, D., van der Meer, A.F.G., van Amersfoort, P.W.: The Free-Electron-Laser User Facility FELIX. Infrared Phys. Technol. 36, 297–308 (1995)
Polfer, N. C.; Oomens, J. Reaction Products in Mass Spectrometry Elucidated with Infrared Spectroscopy. Phys. Chem., Chem. Phys. 9, 3804–3817 (2007)
Valle, J.J., Eyler, J.R., Oomens, J., Moore, D.T., van der Meer, A.F.G., von Helden, G., Meijer, G., Hendrickson, C.L., Marshall, A.G., Blakney, G.T.: Free Electron Laser-Fourier Transform Ion Cyclotron Resonance Mass Spectrometry Facility for Obtaining Infrared Multiphoton Dissociation Spectra of Gaseous Ions. Rev. Sci. Instrum. 76, 7 (2005)
Mize, T.H., Taban, I., Duursma, M., Seynen, M., Konijnenburg, M., Vijftigschild, A., Doornik, C.V., Rooij, G.V., Heeren, R.M.A.: A Modular Data and Control System to Improve Sensitivity, Selectivity, Speed of Analysis, Ease of Use, and Transient Duration in an External Source FTICR-MS. Int. J. Mass Spectrom. 235, 243–253 (2004)
Marshall, A.G., Wang, T.C.L., Ricca, T.L.: Tailored Excitation for Fourier Transform Ion Cyclotron Mass Spectrometry. J. Am. Chem. Soc. 107, 7893–7897 (1985)
Oomens, J., Sartakov, B.G., Meijer, G., von Helden, G.: Gas-Phase Infrared Multiple Photon Dissociation Spectroscopy of Mass-Selected Molecular Ions. Int. J. Mass Spectrom. 254, 1–19 (2006)
Polfer, N.C., Oomens, J.: Vibrational Spectroscopy of Bare and Solvated Ionic Complexes of Biological Relevance. Mass Spectrom. Rev. 28, 468–494 (2009)
Polfer, N.: Infrared Multiple Photon Dissociation Spectroscopy of Trapped Ions. Chem. Soc. Rev. 40, 2211–2221 (2011)
MacAleese, L., Maitre, P.: Infrared Spectroscopy of Organometallic Ions in the Gas Phase: From Model to Real World Complexes. Mass Spectrom. Rev. 26, 583–605 (2007)
Merrill, G.N., Kass, S.R.: Calculated Gas-Phase Acidities Using Density Functional Theory: Is It Reliable? J. Phys. Chem. 100, 17465–17471 (1996)
Steill, J.D., Oomens, J.: Action Spectroscopy of Gas-Phase Carboxylate Anions by Multiple Photon Ir Electron Detachment/Attachment. J. Phys. Chem. A 113, 4941–4946 (2009)
Wabnitz, P.A., Waugh, R.J., Eckersley, M.A., Dua, S., Blumenthal, T., Bowie, J.H.: The Negative Ion Mass Spectra of Deprotonated 2,5-Diketopiperazines. Int. J. Mass Spectrom. Ion Processes 154, 193–201 (1996)
Waugh, R.J., Bowie, J.H., Hayes, R.N.: Collision Induced Dissociations of Deprotonated Peptides: Dipeptides Containing Phenylalanine, Tyrosine. Histidine and Tryptophan. Int. J. Mass Spectrom. Ion Processes 107, 333–347 (1991)
Jones, C.M., Bernier, M., Carson, E., Colyer, K.E., Metz, R., Pawlow, A., Wischow, E.D., Webb, I., Andriole, E.J., Poutsma, J.C.: Gas-Phase Acidities of the 20 Protein Amino Acids. Int. J. Mass Spectrom. 267, 54–62 (2007)
Tian, Z., Wang, X.-B., Wang, L.-S., Kass, S.R.: Are Carboxyl Groups the Most Acidic Sites in Amino Acids? Gas-Phase Acidities, Photoelectron Spectra, and Computations on Tyrosine, p-Hydroxybenzoic Acid, and Their Conjugate Bases. J. Am. Chem. Soc. 131, 1174–1181 (2009)
Oomens, J., Steill, J.D., Redlich, B.: Gas-Phase IR Spectroscopy of Deprotonated Amino Acids. J. Am. Chem. Soc. 131, 4310–4319 (2009)
Sinclair, W.E., Pratt, D.W.: Structure and Vibrational Dynamics of Aniline and Aniline–Ar from High Resolution Electronic Spectroscopy in the Gas Phase. J. Chem. Phys. 105, 15 (1996)
Piest, H.; Helden, v. G.; Meijer, G. Infrared Spectroscopy of Jet-Cooled Neutral and Ionized Aniline-Ar. J. Chem. Phys. 110, 2010–2015 (1999)
Paizs, B.; Suhai, S. Towards Understanding the Tandem Mass Spectra of Protonated Oligopeptides. 1: Mechanism of Amide Bond Cleavage. J. Am. Soc. Mass Spectrom. 15, 103–113 (2004)
Paizs, B.; Suhai, S. Combined Quantum Chemical and RRKM Modeling of the Main Fragmentation Pathways of Protonated GGG. II. Formation of b2, y1, and y2 Ions. Rapid Commun. Mass Spectrom. 16, 375–389 (2002)
Paizs, B., Lendvay, G., Vekey, K., Suhai, S.: Formation of b +2 Ions from Protonated Peptides: An Ab Initio Study. Rapid Commun. Mass Spectrom. 13, 525–533 (1999)
Balta, B., Aviyente, V., Lifshitz, C.: Elimination of Water from the Carboxyl Group of GlyGlyH+. J. Am. Soc. Mass Spectrom. 14, 1192–1203 (2003)
Armentrout, P. B.; Heaton, A. L. Thermodynamics and Mechanisms of Protonated Diglycine Decomposition: A Computational Study. J. Am. Soc. Mass Spectrom., in press (2011).
Acknowledgment
The authors thank Britta Redlich, Lex van der Meer, Giel Berden, and others at the FELIX facility for their skillful assistance. This work is part of the research program of FOM, which is financially supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO). JO thanks the Stichting Physica for support.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic Supplementary Material
Below is the link to the electronic supplementary material.
ESM 1
(PDF 505 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Grzetic, J., Oomens, J. Spectroscopic Evidence for an Oxazolone Structure in Anionic b-Type Peptide Fragments. J. Am. Soc. Mass Spectrom. 23, 290–300 (2012). https://doi.org/10.1007/s13361-011-0297-3
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-011-0297-3