Abstract
Described here is a stable isotope labeling protocol that can be used with a chemical modification- and mass spectrometry-based protein–ligand binding assay for detecting and quantifying both the direct and indirect binding events that result from protein–ligand binding interactions. The protocol utilizes an H 162 O2 and H 182 O2 labeling strategy to evaluate the chemical denaturant dependence of methionine oxidation in proteins both in the presence and absence of a target ligand. The differential denaturant dependence to the oxidation reactions performed in the presence and absence of ligand provides a measure of the protein stability changes that occur as a result of direct interactions of proteins with the target ligand and/or as a result of indirect interactions involving other protein–ligand interactions that are either induced or disrupted by the ligand. The described protocol utilizes the 18O/16O ratio in the oxidized protein samples to quantify the ligand-induced protein stability changes. The ratio is determined using the isotopic distributions observed for the methionine-containing peptides used for protein identification in the LC-MS-based proteomics readout. The strategy is applied to a multi-component protein mixture in this proof-of-principle experiment, which was designed to evaluate the technique’s ability to detect and quantify the direct binding interaction between cyclosporin A and cyclophilin A and to detect the indirect binding interaction between cyclosporin A and calcineurin (i.e., the protein–protein interaction between cyclophilin A and calcineurin that is induced by cyclosporin A binding to cyclophilin A).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Few protein–ligand binding assays can be used for the detection and quantitation of protein–ligand binding interactions in multi-component mixtures, and even fewer techniques exist for analysis of both the direct and indirect binding events that can result from protein–ligand binding interactions. Conventional protein–ligand binding techniques involving spectroscopic or calorimetric methods can only be applied to the analysis of highly purified proteins. While experimental approaches, such as the yeast two-hybrid assay [1–4], have been developed for the large scale and high-throughput analysis of protein–ligand binding interactions, such approaches are generally not quantitative and largely limited to the direct analysis of binary interactions involving two different protein components. Indirect interactions (e.g., those that are induced or precluded as a result of another direct ligand binding interaction) and protein interactions with non-protein ligands are more difficult to assay using the yeast two-hybrid experiments.
Several mass spectrometry-based methods for the analysis of protein–ligand binding interactions have also been developed [5–7]. Mass spectrometry-based proteomic methods used in conjunction with protein complex purification techniques have yielded useful information about protein–protein binding interactions in complex biological mixtures. However, like the yeast two-hybrid binding assay, they have been largely limited to the analysis of protein–protein interactions, and they do not generally yield quantitative information about protein–ligand binding affinities. More recently, amide H/D exchange and covalent labeling methods have been used in combination with mass spectrometry to detect and quantify protein–ligand binding interactions [8–11]. While these approaches have proven useful for the analysis of a wide variety of protein–ligand binding interactions, they are limited to the analysis of ligand binding interactions in relatively simple protein mixtures.
Recently, we reported on a mass spectrometry-based technique that uses a chemical modification strategy, termed stability of proteins from rates of oxidation (SPROX), to detect and quantify protein–ligand binding interactions [12–14]. The methodology involves labeling the methionine side chains of proteins in a hydrogen peroxide-mediated oxidation reaction performed at different chemical denaturant concentrations [12] or increasing temperature [14], both in the presence and absence of a ligand. The extent of oxidation at each denaturant concentration, or temperature, is assessed using a mass spectrometry readout to determine the change in thermodynamic stability imparted to the protein in the presence of the ligand. The SPROX technique is compatible with proteomic fractionation methods such as one- and two-dimensional chromatography, and it can be used to screen complex protein mixtures for ligand binding interactions [13]. An important requirement for SPROX experiments on the proteomic scale is that the extent of protein oxidation must be accurately quantified in the proteomics readout (i.e., the extent of methionine oxidation must be quantified in each of the denaturant-containing SPROX buffers). We recently demonstrated the use of isobaric mass tags for such quantitative measurements using a MudPIT approach [13].
Here we describe the use of a stable isotope labeling strategy termed Probing with SPROX Using Isotope Tags (PrSUIT) for the detection and quantitation of protein–ligand binding interactions by SPROX using a mass spectrometry-based proteomics readout. The strategy involves performing two SPROX analyses on a protein mixture (one on the protein mixture in the absence of ligand and one on the protein mixture in the presence of ligand) using differentially labeled hydrogen peroxide in the oxidation reactions (e.g., H 182 O2 and H 162 O2). Ultimately, a quantitative LC-MS based proteomics readout is used to identify binding proteins (both direct and indirect) of the target ligand. In the proof-of-principle study described here, the PrSUIT technique is applied to a model protein mixture in order to evaluate its ability to detect and quantify the direct binding of the immunosuppressant drug, cyclosporin A (CsA), to yeast cyclophilin A (CPR1), and the indirect binding interaction of CsA with yeast calcineurin that involves the CPR1–CsA complex.
2 Experimental
2.1 Protein Sample Preparation
Saccharomyces cerevisiae overexpression strains (Open Biosystems, Huntsville, AL, USA) in which each yeast open reading frame (ORF) is expressed under control of the GAL1 promoter [15] were used in these experiments. The specific overexpression strains used in this work include those that overexpressed cyclophilin A (CPR1), calcineurin A (CNA1), calcineurin B (CNB1), glycogen synthase (GSY2), pyruvate kinase (PYK1), an uncharacterized ORF (TMA108), and glutamate dehydrogenase (GDH2). The UniProt accession numbers for these yeast proteins were: P14832 (CPR1), P23287 (CNA1), P25296 (CNB1), P40462 (TMA108), P33327 (GDH2), P27472 (GSY2), and P00549 (PYK1). Each of the overexpressed yeast proteins in the overexpression strains also contained a 19 kDa purification tag sequence at its C-terminus. The C-terminal tag contained an HA epitope, a 3C protease cleavage site, a ZZ domain from Protein A, as well as a 6xHis-tag that was exploited in the purification protocol employed here.
Yeast were grown overnight at 30 °C in 2% dextrose SC-Ura, and 2 mL of this solution was used to inoculate 50 mL of 2% raffinose SC-Ura solution, which was then incubated overnight. The solution was diluted into 500 mL of 2% raffinose SC-Ura to give an OD600 of 0.3 and was incubated at 30 °C until the OD600 of the solution was 0.8–1.2. A 250 mL volume of 3×YP solution containing 6% galactose was added to induce overexpression. After 6 h of incubation, yeast cell pellets were obtained by centrifuging 250 mL culture at 1000 × g for 10 min. Cell pellets were lysed in 500 μL of buffer containing 25 mM HEPES, 500 mM NaCl, 10% glycerol, 2 mM mercaptoethanol (pH 7.5), and Halt protease inhibitors (Pierce). Cell lysis was accomplished using 0.5 mm glass beads (Sigma) with 20 s of disruption 10 times and 1 min intervals on ice in between. The samples were then centrifuged at 14,000 × g for 5 min to pellet the insoluble material, the supernatants generated for a given overexpressed protein were combined, and the overexpressed protein was isolated using immobilized metal affinity chromatography (IMAC). The numbers of pellets that were lysed to generate the seven target proteins depended on the overexpression level of each protein and were as follow: 7 pellets for CPR1, 12 pellets for CNA1, 12 pellets for CNB1, 1 pellet for PYK1, 1 pellet for GSY2, 1 pellet for TMA108, and 1 pellet for GDH2.
Talon metal affinity resin (Clontech, Mountain View, CA, USA) was used according to the manufacturer’s directions to isolate the soluble proteins in the supernatants. In these IMAC experiments, the lysate was incubated with the Talon resin for 2 h. The resin was washed three times with 0.5 mL of 50 mM phosphate containing 300 mM NaCl (pH 7), and proteins were eluted off the resin with 450 μL of buffer containing 50 mM phosphate, 300 mM NaCl, and 150 mM imidazole (pH 7). Vivaspin 500 centrifugal filter units (Sartorius Stedim Biotech, Aubagne, France) were used according to the manufacturer’s instructions in order to buffer exchange the IMAC-purified protein samples into a 20 mM phosphate buffer (pH 7.4). The concentration of each IMAC-isolated protein was determined using a Bradford assay [16] in which the total protein concentration was determined. The IMAC-isolated CPR1 and CNA1 samples, which were determined to be 40 and 20 μM, respectively, in total protein, were used directly in the PrSUIT experiments on these two isolated proteins. The model protein mixture used in this work was prepared by combining aliquots of the seven IMAC-isolated proteins (CPR1, CNA1, CNB, PYK1, GDH2, GSY2, and TMA108) into a single mixture. In this mixture, the final concentrations of CPR1, CNA1, CNB, PYK1, GDH2, GSY2, and TMA108 in the mixture were 20, 2, 3, 4, 2, 0.6, and 0.6 μM, respectively. These concentrations were based on the results of our Bradford Assays of the total purified protein concentrations obtained for each IMAC-purified protein.
2.2 PrSUIT Analyses
The denaturant-containing buffers used for the PrSUIT analyses were comprised of 20 mM phosphate buffer (pH 7.4) and guanidinium chloride (GdmCl) (EMD Chemicals, Inc.) concentrations that ranged from 0 to 8 M. The specific [GdmCl] in each buffer was determined by measuring the refractive index of the buffer using the method as described elsewhere [17].
In the PrSUIT experiments, each protein sample (i.e., the model protein mixture, the isolated CPR1 sample, or the isolated CNA1 sample) analyzed here was split into two fractions. An aliquot of a concentrated stock solution of CsA (LKT Laboratories, St. Paul, MN, USA) prepared in DMSO was added to one fraction such that the final concentration of CsA in the protein fraction was 1 mM and the final concentration of DMSO was 10% (vol/vol). An aliquot of DMSO was added to the other fraction such that the final concentration of DMSO in this second fraction (i.e., the sample without ligand) was also 10%. The two fractions (i.e., the one with and the one without ligand) generated for each protein sample analyzed here were equilibrated at room temperature for 30 min. A 20 μL aliquot of each protein sample was combined with 25 μL of each denaturant-containing buffer. The final GdmCl concentrations in the 10 denaturant-containing buffers used in this work were 0.5, 1.0, 1.3, 1.7, 2.0, 2.3, 2.6, 3.0, 3.5, and 4.0 M. The protein samples in each denaturant-containing buffer were equilibrated for 15 min before a 5 μL aliquot of a 0.8 M hydrogen peroxide solution was added to each buffer to initiate the methionine oxidation reaction. The protein samples without CsA were oxidized with H 162 O2 (Sigma) and the protein samples with CsA were oxidized with H 182 O2 (90% enriched; Isotec, Miamisburg, OH, USA). In each case the oxidation reaction in each denaturant-containing buffer was allowed to proceed for 30 min before it was quenched with the addition of 100 μL of a 300 mM methionine (Sigma) solution.
An aqueous solution of TCA (1 g/mL) was added to each denaturant-containing buffer such that the final concentration of TCA was approximately 20% (wt/vol), and the samples were incubated overnight on ice to precipitate the protein. The samples were centrifuged at 8000 × g at 4 °C for 30 min, the supernatants were removed, and the resulting protein pellets were washed three times with 300 μL of ice-cold ethanol. Residual ethanol was removed using a Thermo Savant SpeedVac Concentrator. The protein pellets in each sample were re-dissolved in 60 μL of buffer containing 50 mM ammonium bicarbonate (pH 8.5) and 0.1% Rapigest (Waters, Milford, MA, USA). The samples were heated at 40 °C for 10 min while shaking. Dithiothreitol (Thermo) was added to a final concentration of 10 mM and the sample was heated for 15 min at 80 °C. Iodoacetamide (Thermo) was added to a final concentration of 20 mM and the protein samples were incubated at room temperature for 30 min in the dark. A total of 0.5 μg of trypsin (Sigma) was added to give an enzyme:protein ratio of approximately 1:50, and the samples were incubated overnight at 37 °C while shaking. The trypsin digestion reaction was quenched upon addition of TFA (Halocarbon, River Edge, NJ, USA) and acetonitrile (ACN) such that the final concentrations of TFA and ACN were 1% and 2%, respectively.
The samples were heated at 60 °C for 2 h to cleave the Rapigest, and the peptides were subjected to a second round of oxidation. In this second round of oxidation, a 5 μL aliquot of 0.8 M H 182 O2 was added to the protein samples without ligand and a 5 μL aliquot of 0.8 M H 162 O2 was added to the protein samples with ligand. The second oxidation reaction was allowed to proceed for 2 h before the samples were evaporated to dryness using the SpeedVac Concentrator. The samples were re-dissolved in 50 μL 98:2:0.1 H2O/ACN/FA. After centrifuging each sample at 15,000 × g for 5 min, the samples from the same denaturant-containing buffers but from the with and without ligand fractions were combined to give a set of 10 peptide-containing samples for each PrSUIT analysis.
2.3 LC-MS Analyses
The set of 10 peptide-containing samples generated in each PrSUIT analysis was analyzed using an Agilent 6520 Q-TOF mass spectrometer system equipped with a Chip Cube Interface. The HPLC Chip used in this experiment contained a 43 mm × 75 μm column with Zorbax 300SB-C18 5 μm packing. The tryptic peptides were eluted using a linear gradient from 5% to 50% Buffer B over 30 min and then to 80% Buffer B over 4 min. Buffer A was 0.1% FA in water and Buffer B was 0.1% FA in ACN. The flow rate was 0.4 μL/min. The capillary voltage was 1800 V. The drying gas was 350 °C at a flow rate of 6 L/min. The skimmer and fragmentor were set to 65 and 175 V, respectively. The collision energy was 3.5 V/100 Da with a -4.8 V offset. The inclusion window width for precursor ions was 4 m/z. The scan rate was three scans per s in the mass spectra and two scans per s in the product ion mass spectra, and there were four precursors selected for fragmentation per cycle.
The peptide sequences and proteins in this work were initially identified in five MS/MS analyses performed on a sample composed of 15 μL aliquots from the set of 10 samples generated in the PrSUIT analysis of the model protein mixture. For these five MS/MS analyses a total of 20 μL of the combined sample was loaded on column, which corresponds to an estimated 2–4 μg of total protein, assuming the protein recovery from the protein precipitation step in the PrSUIT protocol was 25%–75%. The methionine-containing peptides analyzed in this work were identified from the product ion mass spectra using Agilent’s Spectrum Mill MS Proteomics Workbench software, Rev A03.03.084 SR4. In the Spectrum Mill searches, the precursor and product ion mass tolerances were set to 20 ppm, the protein cleavage chemistry was set for trypsin with three maximum missed cleavages, and the maximum ambiguous precursor charge was set to 3. Carbamidomethylation of cysteine and oxidation of methionine were set as fixed modifications, and deamidation was set as a variable modification. The peptide fragment products and peptide precursor masses were searched against the NCBInr database for S. cerevisiae. Separate Spectrum Mill searches were also performed to specifically identify peptide sequences with oxidation and dioxidation of cysteine and methionine residues (respectively). In these searches no such peptide sequences were identified with Spectrum Mill scores > 8, which was the minimum score of the peptides identified in our other Spectrum Mill searches.
Agilent’s Bioconfirm was also used to identify some methionine-containing peptides. This software takes the protein’s sequence and searches the mass spectra for tryptic peptides with the expected m/z and charge state. Only methionine-containing peptides that were not identified in the MS/MS runs were inspected, and these were validated based on the appearance of 18O peaks in the isotopic distribution. Extracted ion chromatograms based on the monoisotopic ion signal of each identified methionine-containing peptide (identified with both Spectrum Mill and Bioconfirm) were generated using Agilent’s Qualitative Analysis software.
Ultimately, the %18O labeling of each methionine-containing peptide was determined from the data in a single LC-MS analysis of each of the 10 peptide-containing samples generated in each PrSUIT analysis. The chromatographic conditions used for these LC-MS runs were identical to those described above for the LC-MS/MS runs. A total of 20–30 μL of each peptide-containing sample was loaded on column for these LC-MS runs. This corresponded to ~2–4 μg of total protein for the model protein mixture analysis, and 1.25 and 2 μg for the isolated protein analyses of CPRI and CNA1 (respectively), again assuming sample losses in the protein precipitation and re-dissolution steps in the above PrSUIT protocol were between 25%–75%.
2.4 Calculation of %18O Labeling
Extracted ion chromatograms were generated for each methionine-containing peptide analyzed in this work. The ion signals from the isotopologues detected in the top ~50% of the peak observed in the extracted ion chromatogram for a given methionine-containing peptide were used to calculate an experimentally derived weighted average molecular weight for each methionine-containing peptide. A theoretically derived weighted average molecular weight of each oxidized methionine-containing peptide containing no 18O enrichment was also determined using the natural abundance of each element in the peptide. In the case of peptides containing one methionine residue, the theoretically derived value was subtracted from the experimentally derived value to give a number between 0 and 2, which corresponded to between 0 and 100% 18O labeling (respectively) of the methionine-containing peptides in the PrSUIT protocol. In the case of peptides containing two methionines, the same procedure was applied, except that the theoretically derived value was subtracted from the experimentally derived value to give a number between 0 and 4, which corresponded to between 0 and 100% 18O labeling, respectively. This treatment of multiple methionine residues assumes that the multiple methionine residues in a given peptide have the same level of global protection in the protein’s three-dimensional structure.
The %18O labeling for each peptide in the model mixture analysis was normalized to account for buffer variations (e.g., different TCA precipitation and re-dissolution efficiencies in the different samples). For this normalization, the %18O labeling for all the methionine-containing peptide generated at a specific denaturant concentration were averaged, and the average %18O labeling at each denaturant concentration was divided by the average %18O labeling at the 4 M denaturant concentration to get a set of normalization factors for each denaturant concentration. These normalization factors were 0.68, 0.70, 0.76, 0.78, 0.78, 0.79, 0.81, 0.87, 0.93, and 1.00 for the 0.5, 1.0, 1.3, 1.7, 2.0, 2.3, 2.6, 3.0, 3.5, and 4.0 M denaturant concentrations (respectively). The %18O labeling of each methionine-containing peptide at each denaturant concentration was divided by the specific normalization value at that denaturant concentration to yield a normalized %18O labeling value.
2.5 Construction of SPROX Curves from PrSUIT Plots of Peptide Hits
The PrSUIT plot of a peptide “hit” (i.e., a peptide derived from a protein with a stability change in the presence of ligand) was divided into two sections, one section comprising the data at and below the denaturant concentration of the lowest %18O value in the plot and a second section comprising the data at and above the denaturant concentration of the lowest %18O value in the plot. The %18O values from the first section were used to construct the SPROX curve transition expected in the absence of CsA, and those values from the second section were used to construct the SPROX curve transition in the presence of CsA. The transitions of the SPROX curves (i.e., Fraction Oxidized values versus [Denaturant] plots) expected with and without ligand were constructed with Fraction Oxidized values that were calculated using Equations 1 and 2, respectively.
In Equations 1 and 2, %18O is the normalized %18O value at each denaturant concentration, %18Olow is the lowest %18O value observed for the peptide, and %18Oave is the average %18O value observed for all the methionine-containing peptides, which was 39% in this work.
2.6 Binding Affinity Measurements
The K d values for the CPR1-CsA complex and for the CPR1-CsA-CNA1 complex were determined using Equation 3.
In Equation 3, [L] is the concentration of free ligand, N is the number of independent equivalent binding sites, ∆∆Gf is the binding free energy, R is the ideal gas constant, and T is the temperature. In these experiments N = 1 and the concentration of free ligand was estimated as the total ligand concentration, as both the CsA and CPR1-CsA ligands in this work were present in a large excess over their protein targets. The free CsA and CPR1-CsA concentrations were estimated as 400 and 8 μM (respectively), with their protein targets, CPR1 and CNA1 (respectively), having estimated concentrations of 8 and 0.8 μM (respectively).
The ∆∆Gf values used in our binding affinity measurements were calculated from ∆Gf values derived from the SPROX curve transitions constructed from the PrSUIT data on the CPR1(54-74) and CNA1(332-354) peptide “hits.” The ∆Gf values were obtained by fitting the SPROX curve transitions (i.e., Fraction Oxidized versus [Denaturant] plots) to Equation 4.
In Equation 4, Kfold = e-(∆Gf+m[Den])/RT, FrOx0 is the fraction oxidized before the first oxidation reaction in PrSUIT, FrOx∞ is fraction oxidized after the second oxidation reaction in PrSUIT, t is the time of the first oxidation in seconds, kOX is the average pseudo-first order rate constant for the oxidation of an unprotected methionine residue in the protein, ∆Gf is the free energy of folding in the absence of denaturant, [Den] is the denaturant concentration, m is δ∆Gf/δ[GdmCl], R is the gas constant, and T is the temperature in Kelvin. In fitting the Fraction Oxidized versus [Denaturant] plots to Equation 3, the FrOx0 and FrOx∞ were assigned values of 0 and 1 (respectively), ∆Gf, and m were allowed to float, and kOX was assigned a value of 4.1 hr–1, which was based on the 80 mM H2O2 concentration used in the first oxidation reaction and the second order rate constant for the oxidation of unprotected methionine residues in proteins that we previously determined to be approximately 50.4 M–1 hr–1. Equation 4 is identical to the equation we previously described in reference 12 to analyze SPROX data (i.e., Equation 2 in reference 12) with the exception that the ∆Masswt,av, ∆M∞, and ∆M0 were replaced with Fraction Oxidized, FrOx∞, and FrOx0 (respectively).
3 Results
3.1 General Protocol
The stable isotope labeling strategy developed in this work is outlined in Figure 1. In this strategy, hydrogen peroxide isotopically enriched with 18O (H 182 O2) and hydrogen peroxide containing naturally occurring 16O (H 162 O2) are used to selectively oxidize the methionine residues in a protein sample pre-equilibrated with and without ligand (respectively) at a series of different chemical denaturant concentrations. The oxidation reactions are quenched and the protein sample in each denaturant-containing buffer is subjected to an LC-MS-based proteomics analysis using a bottom-up approach that involves generating tryptic peptides of the proteins from each denaturant containing buffer. Prior to the LC-MS readout, the tryptic peptides from the proteins in each denaturant-containing buffer are subjected to a second round of oxidation to quantitatively oxidize all of the methionine side chains in the samples to methionine sulfoxide.
This second round of oxidation oxidizes the methionine side chains that were globally protected (i.e., buried in a protein’s folded three-dimensional structure) and not oxidized during the first round of oxidation. The second round oxidation reaction conditions are tuned such that the methionine sulfoxide formed in the first round oxidation reactions are not further oxidized to the sulfone. The second round oxidation reactions also involve the use of H 182 O2 and H 162 O2. However, in the second round of oxidation, the peptide mixtures derived from the protein samples originally incubated with and without ligand are now reacted with H 162 O2 and H 182 O2, respectively (i.e., the opposite of that done in the first round). This creates isotopic diversity between the methionine-containing peptides derived from the protein samples that were originally in the presence and absence of ligand, and permits the samples prepared with and without ligand to be paired and combined (see Figure 1) such that they can be simultaneously analyzed in the same LC-MS run.
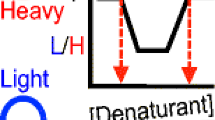
Ultimately, the chemical denaturant dependence of the 18O enrichments observed for the methionine-containing peptides detected in the LC-MS readout are used to detect and quantify binding interactions of the target ligand with different proteins in the mixture (Figure 2). Proteins that do not interact with the target ligand will have the same 18O enrichment at all denaturant concentrations. This is because the chemical denaturant dependence observed in the first round of oxidation reactions performed in the presence and absence of ligand will be the same for proteins that do not have their thermodynamic stability modulated through interaction with the ligand (see “no interaction” SPROX curves in Figure 2a). However, proteins that do have their thermodynamic stability modulated through interaction with the ligand will have a denaturant dependence to their oxidation in the first round of oxidation that is different in the presence and absence of ligand (see “interaction” SPROX curves in Figure 2a). The 18O enrichment observed for methionine-containing peptides derived from regions of protein structure that experience different degrees of global and/or subglobal protection upon ligand binding will be more (or less) enriched with 18O at one or more denaturant concentrations (see Figure 2a and b).
Theoretical structure of PrSUIT data and its relationship to SPROX data. (a) Hypothetical PrSUIT data (i.e., %18O versus [Den] plots) expected for proteins that do not (Case I) and do (Cases II or III) have their thermodynamic stability altered in the presence of the ligand. Cases II and III correspond to situations in which the thermodynamic stability of the identified protein is stabilized and destabilized (respectively) as a result of ligand binding. (b) Schematic representation of the molecular 16O and 18O oxidized species generated in the first and second oxidation reactions in the PrSUIT protocol at three different points in a protein’s SPROX curves generated in the absence, (–), and in the presence, (+), of a binding ligand
3.2 PrSUIT Analysis of a Model Protein Mixture
The protocol outlined in Figure 1 was used to characterize the binding of CsA to the individual proteins in a model protein mixture, which was mainly composed of seven yeast proteins including: CPR1, CNA1, CNB1, PYK1, GDH2, TMA108, and GSY2. The mixture also contained a number of additional yeast proteins that were co-purified in the IMAC step used to isolate the above seven proteins from yeast cell lysates in which they were each overexpressed. The primary goal of this work was to determine if the known direct and indirect interactions of CsA with CPR1 and CNA1 (respectively) could be detected using the stable isotope labeling protocol outlined here.
Initially, a pooled sample containing equal aliquots of the 10 combined sample pairs (see Figure 1) was subjected to an LC-MS/MS analysis to sequence the peptides and identify the proteins present in the sample. A total of 120 peptides from 49 different proteins were identified using Spectrum Mill to search the mass spectral data obtained in five LC-MS/MS runs. These Spectrum Mill identifications included 29 methionine-containing peptides from a total of 21 different proteins and 13 methionine-containing peptides from six of the seven primary proteins in the mixture. While CNB1 was successfully identified as a component of the mixture, its identification was not made with a methionine-containing peptide. An additional seven methionine-containing peptides from the seven primary proteins in the mixture were also identified based on their expected mass and isotope envelope using Agilent’s Bioconfirm software. The sequences of these additional seven methionine-containing peptides were subsequently confirmed in LC-MS/MS experiments specifically targeting these peptides. Ultimately, 36 methionine-containing peptides from 21 different proteins were analyzed in the PrSUIT analysis described here (see Table 1).
The 10 paired and combined samples were each subjected to an LC-MS analysis, and the LC-MS data (i.e., the relative intensities of the isotopologues detected in the spectra obtained from the methionine-containing peptides) was used to determine the %18O incorporation in each methionine-containing peptide at each denaturant concentration (see Figure 3). The resulting %18O incorporation values were then normalized to correct for systematic errors that could result from either differential protein recoveries and/or differences in the actual H 162 O2 and H 182 O2 concentrations. Such values for 27 of the 36 methionine-containing peptides identified in the model mixture could be readily determined. The normalized %18O labeling values for nine of the methionine-containing peptides identified in the model mixture were compromised because the peptides had low intensity ion signals and/or ion signals that overlapped with those of other co-eluting peptides.
Relative ion signal intensities observed (black bars) and theoretically calculated (grey bars) for the isotopologues of the methionine-containing peptide, CPR1(54-74) of sequence VIPDFMLQGGDFTAGNGTGGK, that was detected in the PrSUIT analysis of the model protein mixture. The data obtained at two different denaturant concentrations, 3.5 and 2.0 M GdmCl, are shown in (a) and (b), respectively. In each case M represents the monoisotopic mass of the peptide, 2096.9837 Da. The theoretical distributions of the relative ion signal intensities for the isotopologues in (a) and (b) are those expected for the oxidized CPR1(54-74) peptide with 41 and 11% 18O labeling, respectively, in the PrSUIT protocol
PrSUIT plots (i.e., Normalized %18O Labeling versus [GdmCl] plots) were constructed for 27 methionine-containing peptides detected in the model mixture analysis (see Table 1). These 27 methionine-containing peptides included 20 peptides from six of the seven target proteins in this study as well as seven peptides from seven other random proteins that were apparently co-purified with our seven target proteins. The seven peptides from these seven random proteins in the mixture served as negative controls. Shown in Figure 4 are PrSUIT plots for several methionine-containing peptides, including one with a relatively constant %18O labeling at the different denaturant concentrations (Figure 4a) and two with %18O labeling that is significantly low (see below) at several denaturant concentrations (Figure 4b and c).
Representative PrSUIT results obtained in the CsA binding analysis using the model protein mixture (closed symbols) and the purified proteins (open symbols). (a) PrSUIT plot obtained for a methionine-containing peptide, GDH2(920-943) of sequence GGVTSSSMEVLASLALNDNDFVHK, derived from a protein with no CsA-induced interactions. (b) and (c) PrSUIT plots obtained for two methionine-containing peptides, CPR1 (54-74) of sequence VIPDFMLQGGDFTAGNGTGGK and CNA1(332-354) of sequence VTGFPSLITMFSAPNYLDTYHNK (respectively), each derived from proteins with known CsA-induced interactions. The CsA binding interaction with CNA1 is also known to require CPR1. Note that the purified protein plots were constructed with %18O labeling values that were not normalized
Shown in Figure 5 is the distribution of normalized %18O labeling values obtained at all the denaturant concentrations for all of the 27 methionine-containing peptides in this work. Also shown in Figure 5 is the distribution of normalized %18O labeling values obtained for just the negative controls. Both distributions are similar, although the distribution observed for the negative controls is not as smooth due to the relatively small number of points. The range of normalized %18O labeling values observed in our experiments was largely defined by the random error associated with our weighted mass average determinations for each peptide. The largest source of error in the weighted mass average determinations came from the variability associated with the relative ion signal intensities of the different isotopologues of a given peptide. It is this error that largely defined the 6% standard deviation observed for all the normalized %18O labeling values in Figure 5.
Distribution of all the normalized %18O labeling values at all the denaturant concentrations (black bars) obtained on the 27 methionine-containing peptides analyzed in the PrSUIT experiment performed on the model protein mixture, as well as the contribution of normalized %18O values from just the negative control peptides (white bars)
The distribution of all the normalized %18O labeling values for all the peptides in Figure 5 is also centered at the average value, 39%, which is close to the 45% value expected for methionine-containing peptides derived from proteins that do not interact with CsA. The %18O incorporation into methionine-containing peptides from proteins that do not interact with CsA is expected to be 45% if the H 182 O2 reagent was indeed 90% enriched with 18O as reported by the supplier, and if the H 182 O2 and H 162 O2 reagent concentrations in the first round oxidation reactions were indeed identical. The 6% discrepancy between the average %18O value observed in our experiment, 39%, and the expected value, 45%, is most likely due to the H2O 182 and H2O 162 reagent concentrations not being exactly identical during the first round of oxidation. Our results suggest that the actual hydrogen peroxide concentration in the H 182 O2 stock solution may have been slightly lower than that in the H 162 O2 stock solution. Hydrogen peroxide solutions can decompose over time to water and oxygen, thus it is possible that the actual hydrogen peroxide concentrations in the H 182 O2 and H 162 O2 stock solutions may have been differentially lower than that reported by the manufacturer. Any oxidization of the protein material in the samples prior to PrSUIT analysis (i.e., oxidation products generated during the protein purification steps) could also have contributed to the systematic reduction in 18O levels observed in our PrSUIT analyses.
The distribution of all the normalized %18O labeling values at all the denaturant concentrations (see Figure 5) was used to help identify peptides with significantly altered %18O labeling. Methionine-containing peptides with normalized %O18 labeling greater than 51% or less than 27% were deemed significantly altered based on the observed distribution of the measured values (Figure 5), which revealed 96% of the measured values were within the 27-51% range. The center of this range, 39%, was the average normalized %O18 labeling value determined for all the methionine-containing peptides at all the denaturant concentrations. The range was also equivalent to the average value ±2 standard deviations. We also reasoned that tight binding interactions would produce such altered normalized %18O labeling values that were consistently greater than 51% or consistently less than 27% at two or more consecutive denaturant concentrations. This selection strategy is analogous to that which we have previously described for selecting tight binding interactions in SPROX analyses using an isobaric mass tagging strategy [13].
A total of 24 of the 27 methionine-containing peptides analyzed here had normalized %18O labeling values between 27% and 51% at all the denaturant concentrations and were clearly not “hits.” One peptide had normalized %18O labeling values outside the 27% and 51% at only one denaturant concentration and was also not categorized as a “hit.” Two methionine-containing peptides, including one derived from CPR1 and one derived from CNA1 were identified as “hits” as they had normalized %18O labeling values that were less than 27% at two or more consecutive denaturant concentrations. The methionine-containing peptide from CPR1, CPR1(54-74) of sequence VIPDFMLQGGDFTAGNGTGGK, had normalized %18O labeling values less than 27% at five denaturant concentrations between 1 M and 3 M GdmCl (see Figure 4b), indicating an increase in thermodynamic stability of the CPR1 protein in the presence of CsA. The CNA1 peptide, CNA1(332-354) of sequence VTGFPSLITMFSAPNYLDTYHNK, had normalized %18O labeling values less than 27% at two consecutive denaturant concentrations between 2 M to 3 M (Figure 4c), indicating an increase in thermodynamic stability of the CNA1 protein in the presence of CsA.
3.3 PrSUIT Analyses of Purified CPR1 and Purified CNA1
As part of this work, IMAC-purified samples of overexpressed CPR1 and CNA1 were each subjected to a PrSUIT experiment using the CsA ligand. The single methionine-containing peptide of CPR1 (i.e., CPR1(54-74)) that was detected in the PrSUIT analysis of CPR1 in the model protein mixture was also the single methionine-containing CPR1 peptide detected in the PrSUIT analysis of the purified protein. The PrSUIT plot obtained for the CPR1(54-74) peptide in the experiment on the purified CPR1 protein (see Figure 4b, open symbols) had the same overall structure as the PrSUIT plot obtained for the peptide in the experiment performed on the model protein mixture (Figure 4b, closed symbols), although the exact %18O labeling values recorded in the two experiments were not identical. Both plots show similarly reduced 18O labeling at multiple denaturant concentrations between 1 M and 3 M GdmCl (see Figure 4b), indicating an increase in thermodynamic stability of the CPR1 protein in both PrSUIT experiments, a result that is consistent with CPR1 being a direct protein target of CsA. The discrepancies between the %18O labeling values recorded in the two experiments can be explained by small differences in the experimental parameters (e.g., mismatched H 182 O2 and H 162 O2 reagent concentrations) and/or potential differences in the biophysical properties of the CPR1 folding reaction in the presence of CNA1 and CNB (see the Discussion section).
Seven unique methionine-containing peptides from CNA1 were identified in the PrSUIT experiment performed on the purified CNA1 construct (see Table 1). The seven methionine-containing peptides included one CNA1 peptide that was identified as a “hit” in the PrSUIT analysis of the model protein mixture, two CNA1 peptides that were detected in the PrSUIT analysis of the model mixture but not identified as “hits,” and four new methionine-containing CNA1 peptides that were not detected in the PrSUIT analysis of the model protein mixture. The PrSUIT plots obtained for all seven of the methionine-containing CNA1 peptides identified in the experiment on the purified CNA1 construct showed relatively constant 18O incorporation at all denaturant concentrations. Of particular significance is that the PrSUIT plot for the CNA1(332-354) peptide, which had displayed reduced 18O incorporation at multiple denaturant concentrations between 2 M and 3 M GdmCl (see Figure 4c) when CNA1 was analyzed in the model protein mixture, did not yield such reduced 18O incorporation when CNA1 was analyzed separately from the mixture (see Figure 4c). This change in the PrSUIT behavior of the CNA1(332-354) peptide is consistent with CNA1 being an indirect protein target of CsA.
3.4 Binding Affinity Measurements
The PrSUIT data collected on the “hit” peptides identified in our PrSUIT experiments was used to generate SPROX curves (see the Experimental section) for each peptide hit in the presence and absence of ligand. The resulting SPROX curves were used to quantify the affinity of the detected protein–ligand interaction. Shown in Figure 6 are the SPROX curves generated from the PrSUIT data collected here on the CNA1(332-354) and CPR1(54-74) peptides derived from the protein mixture (see Figure 4b and c). The ∆Gf values extracted from the SPROX curves generated for the CPR1(54-74) peptide with and without the CsA ligand (–12.0 ± 2.6 and –5.5 ± 2.3 kcal/mol, respectively) and the ∆Gf values extracted from the SPROX curves generated for the CNA1(332-354) peptide with and without the CsA ligand (–7.1 ± 2.1 and –4.7 ± 2.2 kcal/mol, respectively) were used to calculate K d values of 6 and 120 nM for the CPR1-CsA and CPR1-CsA-CNA1 complexes, respectively.
SPROX curves generated from PrSUIT data collected in this work on CPR1 and CNA1 in the model mixture analysis. (a) SPROX curves generated for the CPR1(54-74) peptide of sequence VIPDFMLQGGDFTAGNGTGGK using the PrSUIT data shown in Figure 4b. (b) SPROX curves generated for the CNA1(332-354) peptide of sequence VTGFPSLITMFSAPNYLDTYHNK using the PrSUIT data shown in Figure 4c
4 Discussion
The primary goal of this work was to determine if the stable isotope labeling protocol outlined here could detect and quantify the direct binding of CsA to CPR1 and the indirect binding interaction of CsA with calcineurin that involves the CPR1-CsA complex. The binding affinity of CsA for human cyclophilin A has been previously measured using more conventional techniques, and dissociation constants in the 30–200 nM range have been reported [18–22]. In our earlier SPROX experiments on cyclophilin A, including our work on the intact human protein using a MALDI readout [12] and on the yeast protein using a quantitative proteomics readout involving a isobaric mass tagging strategy [13], the K d values determined for the CsA-cyclophilin A complex ranged from 26 to 100 nM. The K d value determined here from our PrSUIT data, 6 nM, is close to this range (i.e., within 5-fold of the lower end).
The interaction of calcineurin with the CsA-cyclophilin A complex has been established in enzymatic [23, 24] and X-ray crystallographic studies [25], and the interaction has been shown in these studies to require both cyclophilin and CsA (i.e., neither cyclophilin nor CsA interact with calcineurin in the absence of the other). Our PrSUIT results on the yeast proteins in this study are also consistent with this observation as the CNA1(332-354) peptide was not identified as a “hit” in our PrSUIT experiment on the isolated CNA1 protein. It was, however, identified as a “hit” in the protein mixture analysis, in which CNA1 was analyzed in the presence of CPR1. To our knowledge a K d value has not been previously reported in the literature for the calcineurin binding interaction with the CsA-cyclophilin A complex. However, the human cyclophilin A-CsA complex has been shown to inhibit the phosphatase activity of calcineurin with a KI value of 32 nM [23], which is within 5-fold of the K d value of 120 nM that was determined here.
Five of the proteins in the model mixture were identified to interact (either directly and/or indirectly) with CsA in our earlier work in which the binding assay was performed in the context of all the endogenous proteins in a yeast cell lysate [13]. Peptide hits from these proteins were not identified in the PrSUIT experiment described here. These results suggest that CsA does not directly interact with these proteins. Our identification of these proteins as hits in our earlier analyses of these proteins in the context of the yeast cell lysate suggests that there are other yeast proteins that mediate CsA’s interaction with these proteins.
The PrSUIT strategy described here is fundamentally related to SPROX. Thus, as is the case for SPROX analyses, a requirement for PrSUIT analyses is that the protein(s) under study must have methionine residues in their primary amino acid sequence that are globally protected in the protein’s folded three-dimensional structure. Such globally protected methionine residues are methionine residues that can only react with the hydrogen peroxide reagent when the protein globally unfolds. The successful analyses of such proteins using the LC-MS-based proteomics readout described here further requires that tryptic peptides containing these methionine residues also be detected in the mass spectrometry readout. The results obtained in this work and in our earlier SPROX study indicate that approximately 20%–25% of the peptides identified in the LC-MS-based readout contained at least one methionine residue and these peptides mapped to between 30% and 45% of the identified proteins.
Of particular significance in both SPROX and PrSUIT analyses of protein–ligand binding is that the methionine residues need not be derived from regions of a protein’s three-dimensional structure that are at or near the binding site. They can be derived from any globally protected region of the folding domain(s) in the protein that are involved in binding. This is substantiated by our PrSUIT results, which identified both the CNA1(332-354) and CPR1(54-74) peptides as “hits.” These yeast CNA1 and CPR1 peptide sequences can be aligned in human homologues of CNA1 and CPR1, on which there is X-ray crystallographic data for the CsA-CPR1 and CsA-CPR1-CNA1 complexes, and it can be determined that the CNA1(332-352) peptide is at the CPR1-CNA1 binding interface, but that the CPR1(54-74) peptide is not at the CPR1-CsA binding interface.
As with SPROX, there are several caveats for the use of PrSUIT to detect and quantify protein–ligand binding interactions. The determination of K d values requires an accurate evaluation of the free ligand concentration (see Equation 3). The successful detection of a protein–ligand binding interaction using either SPROX or PrSUIT also requires a sufficiently high free ligand concentration to produce a measurable shift (i.e., >0.5 M GdmCl) in the protein’s SPROX curve. The minimum free ligand concentration needed to produce a >0.5 M shift will not be the same for all protein–ligand systems. Complexes with larger K d values (i.e., weaker binding interactions) and larger m-values (i.e., more cooperative folding reactions) will require higher free ligand concentrations for their detection than those with smaller K d values and smaller m-values. Our results with the CPR1 and CNA1 protein systems suggest that nanomolar K d values should be readily measured using free ligand concentrations in the 10–400 μM range for proteins with m-values in the 2–4 kcal/(mol M), respectively.
In SPROX and PrSUIT analyses of protein–ligand binding, it is also important that the oxidation products of H2O2-mediated oxidation reaction do not alter the ligand binding affinities of the protein(s) under study. The reasonably good agreement between the PrSUIT derived binding affinities determined in this work and those previously reported for the CsA-CPR1 and CsA-CPR1-CNA1 complexes suggest that the oxidation products did not complicate the analyses described here. It is also important that the protein–ligand binding affinity not be compromised by the presence of the chemical denaturant.
The basic structure of PrSUIT data is expected to be like what is summarized in Figure 2a. However, the specific %18O labeling values observed in a PrSUIT experiment will depend on several experimental parameters and on the biophysical properties of the protein–ligand interaction under study. For example, the baseline %18O labeling values observed in a PrSUIT analysis will be maximized when the 18O enrichment of the H 182 O2 reagent is high, the H 182 O2 and H 162 O2 reagent concentrations are exactly matched, and “air” oxidation of the sample prior to analysis is minimized. The magnitude of the “dip” in %18O labeling values expected for binding proteins will be greatest when the baseline %18O labeling values are maximized. The exact magnitude of the “dip” in %18O labeling values expected for binding proteins will also vary from protein complex to protein complex. The magnitude of the “dip” is largely dependent on the amplitudes of the SPROX curves expected for the protein and the protein–ligand complex. The fraction oxidized values in the post-transition baselines of SPROX curves (see Figure 2) are typically 1. However, fraction oxidized values in the pre-transition baselines of SPROX curves can vary depending on how well a specific methionine residue is protected from oxidation in the SPROX experiment. Methionine residues that are buried in the hydrophobic core of a protein (or protein–ligand complex) and that are in conformationally restricted regions of the protein’s folded three-dimensional structure will be more highly protected from oxidation in the SPROX experiment and will yield SPROX curves with low pre-transition baselines and high amplitudes. However, globally protected methionine residues in more dynamic (i.e., conformationally flexible) regions of protein structure can yield SPROX curves with larger pre-transition baseline values and small amplitudes.
In the PrSUIT experiment, it is important that the same protein concentration be used in the plus and minus ligand samples and that there is a consistent level of protein recovery in the PrSUIT sample workup (particularly in the protein precipitation and re-dissolution steps). Differences in protein concentrations will skew the %18O values observed in the baselines of the PrSUIT plots. It is relatively straightforward to ensure that the same protein concentration is used for the plus and minus ligand samples by employing the same protein stock solution to prepare both sets of samples. However, it can be difficult to ensure a consistent level of protein recovery in the PrSUIT sample workup. For example, the protein precipitation and re-dissolution steps can yield different amounts of protein from the chemical denaturant-containing buffers. If the protein recovery is not consistent in the samples that are paired prior to the LC-MS/MS analysis, the resulting %18O values will be skewed. In ligand binding analyses involving protein mixtures where few protein–ligand binding interactions are expected (i.e., the majority of methionine-containing peptides are expected to exhibit non-binding behavior shown in Figure 2a), it is possible to correct for such sample-to-sample variation in protein recoveries using the normalization procedure described above.
The PrSUIT strategy developed here for the detection and quantitation of protein ligand binding interactions by SPROX has several advantages over the isobaric mass tagging strategy we previously reported. The labeling scheme in PrSUIT generates only one form (i.e., the sulfoxide form) of all the methionine-containing peptides in a sample, and it enables quantitative data to be extracted from the mass spectral data. This is in contrast to the isobaric mass tagging strategy we previously demonstrated with SPROX, which generates two forms (i.e., oxidized and non-oxidized) of each methionine-containing peptide in the samples, and requires product ion mass spectra for quantitation. The reduction in sample complexity that comes with generation of only the oxidized form of methionine-containing peptides in PrSUIT may facilitate the analysis of more complex protein mixtures. The ability to extract quantitative data from mass spectral data may also facilitate more complete coverage of the methionine-containing peptides generated in the PrSUIT analysis of proteins in complex protein mixtures.
References
Uetz, P., Giot, L., Cagney, G., Mansfield, T.A., Judson, R.S., Knight, J.R., Lockshon, D., Narayan, V., Srinivasan, M., Pochart, P., Qureshi-Emili, A., Li, Y., Godwin, B., Conover, D., Kalbfleisch, T., Vijayadamodar, G., Yang, M.J., Johnston, M., Fields, S., Rothberg, J.M.: A comprehensive analysis of protein–protein interactions in Saccharomyces cerevisiae. Nature 403, 623–627 (2000)
Giot, L., Bader, J.S., Brouwer, C., Chaudhuri, A., Kuang, B., Li, Y., Hao, Y.L., Ooi, C.E., Godwin, B., Vitols, E., Vijayadamodar, G., Pochart, P., Machineni, H., Welsh, M., Kong, Y., Zerhusen, B., Malcolm, R., Varrone, Z., Collis, A., Minto, M., Burgess, S., McDaniel, L., Stimpson, E., Spriggs, F., Williams, J., Neurath, K., Ioime, N., Agee, M., Voss, E., Furtak, K., Renzulli, R., Aanensen, N., Carrolla, S., Bickelhaupt, E., Lazovatsky, Y., DaSilva, A., Zhong, J., Stanyon, C.A., Finley Jr., R.L., White, K.P., Braverman, M., Jarvie, T., Gold, S., Leach, M., Knight, J., Shimkets, R.A., McKenna, M.P., Chant, J., Rothberg, J.M.: A protein interaction map of Drosophila melanogaster. Science 302, 1727–1736 (2003)
Li, S.M., Armstrong, C.M., Bertin, N., Ge, H., Milstein, S., Boxem, M., Vidalain, P.O., Han, J.D.J., Chesneau, A., Hao, T., Goldberg, D.S., Li, N., Martinez, M., Rual, J.F., Lamesch, P., Xu, L., Tewari, M., Wong, S.L., Zhang, L.V., Berriz, G.F., Jacotot, L., Vaglio, P., Reboul, J., Hirozane-Kishikawa, T., Li, Q.R., Gabel, H.W., Elewa, A., Baumgartner, B., Rose, D.J., Yu, H.Y., Bosak, S., Sequerra, R., Fraser, A., Mango, S.E., Saxton, W.M., Strome, S., van den Heuvel, S., Piano, F., Vandenhaute, J., Sardet, C., Gerstein, M., Doucette-Stamm, L., Gunsalus, K.C., Harper, J.W., Cusick, M.E., Roth, F.P., Hill, D.E., Vidal, M.: A map of the interactome network of the metazoan C. elegans. Science 303, 540–543 (2004)
Rual, J.F., Venkatesan, K., Hao, T., Hirozane-Kishikawa, T., Dricot, A., Li, N., Berriz, G.F., Gibbons, F.D., Dreze, M., Ayivi-Guedehoussou, N., Klitgord, N., Simon, C., Boxem, M., Milstein, S., Rosenberg, J., Goldberg, D.S., Zhang, L.V., Wong, S.L., Franklin, G., Li, S.M., Albala, J.S., Lim, J.H., Fraughton, C., Llamosas, E., Cevik, S., Bex, C., Lamesch, P., Sikorski, R.S., Vandenhaute, J., Zoghbi, H.Y., Smolyar, A., Bosak, S., Sequerra, R., Doucette-Stamm, L., Cusick, M.E., Hill, D.E., Roth, F.P., Vidal, M.: Towards a proteome-scale map of the human protein–protein interaction network. Nature 437, 1173–1178 (2005)
Ho, Y., Gruhler, A., Heilbut, A., Bader, G.D., Moore, L., Adams, S.L., Millar, A., Taylor, P., Bennett, K., Boutilier, K., Yang, L.Y., Wolting, C., Donaldson, I., Schandorff, S., Shewnarane, J., Vo, M., Taggart, J., Goudreault, M., Muskat, B., Alfarano, C., Dewar, D., Lin, Z., Michalickova, K., Willems, A.R., Sassi, H., Nielsen, P.A., Rasmussen, K.J., Andersen, J.R., Johansen, L.E., Hansen, L.H., Jespersen, H., Podtelejnikov, A., Nielsen, E., Crawford, J., Poulsen, V., Sorensen, B.D., Matthiesen, J., Hendrickson, R.C., Gleeson, F., Pawson, T., Moran, M.F., Durocher, D., Mann, M., Hogue, C.W.V., Figeys, D., Tyers, M.: Systematic identification of protein complexes in Saccharomyces cerevisiae by Mass Spectrometry. Nature 415, 180–183 (2002)
Graumann, J., Dunipace, L.A., Seol, J.H., McDonald, W.H., Yates, J.R., Wold, B.J., Deshaies, R.J.: Applicability of tandem affinity purification MudPIT to pathway proteomics in yeast. Mol. Cell. Proteom. 3, 226–237 (2004)
Gavin, A.C., Bosche, M., Krause, R., Grandi, P., Marzioch, M., Bauer, A., Schultz, J., Rick, J.M., Michon, A.M., Cruciat, C.M., Remor, M., Hofert, C., Schelder, M., Brajenovic, M., Ruffner, H., Merino, A., Klein, K., Hudak, M., Dickson, D., Rudi, T., Gnau, V., Bauch, A., Bastuck, S., Huhse, B., Leutwein, C., Heurtier, M.A., Copley, R.R., Edelmann, A., Querfurth, E., Rybin, V., Drewes, G., Raida, M., Bouwmeester, T., Bork, P., Seraphin, B., Kuster, B., Neubauer, G., Superti-Furga, G.: Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415, 141–147 (2002)
Powell, K.D., Ghaemmaghami, S., Wang, M.Z., Ma, L.Y., Oas, T.G., Fitzgerald, M.C.: A general mass spectrometry-based assay for the quantitation of protein–ligand binding interactions in solution. J. Am. Chem. Soc. 124, 10256–10257 (2002)
Zhu, M.M., Rempel, D.L., Du, Z.H., Gross, M.L.: Quantification of protein–ligand interactions by mass spectrometry, titration, and H/D exchange: PLIMSTEX. J. Am. Chem. Soc. 125, 5252–5253 (2003)
Charvatova, O., Foley, B.L., Bern, M.W., Sharp, J.S., Orlando, R., Woods, R.J.: Quantifying protein interface footprinting by hydroxyl radical oxidation and molecular dynamics simulation: application to Galectin-1. J. Am. Soc. Mass Spectrom. 19, 1692–1705 (2008)
Takamoto, K., Chance, M.R.: Radiolytic protein footprinting with mass spectrometry to probe the structure of macromolecular complexes. Annu. Rev. Biophys. Biomol. Struct. 35, 251–276 (2006)
West, G.M., Tang, L., Fitzgerald, M.C.: Thermodynamic analysis of protein stability and ligand binding using a chemical modification- and mass spectrometry-based strategy. Anal. Chem. 80, 4175–4185 (2008)
West, G.M., Tucker, C.L., Xu, T., Park, S.K., Han, X.M., Yates, J.R., Fitzgerald, M.C.: Quantitative proteomics approach for identifying protein-drug interactions in complex mixtures using protein stability measurements. Proc. Natl. Acad. Sci. U. S. A. 107, 9078–9082 (2010)
West, G.M., Thompson, J.W., Soderblom, E.J., Dubois, L.G., DeArmond, P.D., Moseley, M.A., Fitzgerald, M.C.: Mass spectrometry-based thermal shift assay for protein–ligand binding analysis. Anal. Chem. 82, 5573–5581 (2010)
Gelperin, D.M., White, M.A., Wilkinson, M.L., Kon, Y., Kung, L.A., Wise, K.J., Lopez-Hoyo, N., Jiang, L.X., Piccirillo, S., Yu, H.Y., Gerstein, M., Dumont, M.E., Phizicky, E.M., Snyder, M., Grayhack, E.J.: Biochemical and genetic analysis of the yeast proteome with a movable ORF collection. Genes Dev. 19, 2816–2826 (2005)
Bradford, M.M.: Rapid and sensitive method for quantitation of microgram quantities of protein utilizing principle of protein-dye binding. Anal. Biochem. 72, 248–254 (1976)
Nozaki, Y.: The preparation of guanidine hydrochloride. Method Enzymol. 26, 43–50 (1972)
Handschumacher, R.E., Harding, M.W., Rice, J., Drugge, R.J.: Cyclophilin—a specific cytosolic binding-protein for cyclosporin A. Science 226, 544–547 (1984)
Harding, M.W., Handschumacher, R.E.: Cyclophilin, a primary molecular target for cyclosporine—structural and functional implications. Transplantation 46, S29–S35 (1988)
Liu, J., Albers, M.W., Chen, C.M., Schreiber, S.L., Walsh, C.T.: Cloning, expression, and purification of human cyclophilin in Escherichia coli and assessment of the catalytic role of cysteines by site-directed mutagenesis. Proc. Natl. Acad. Sci. U. S. A. 87, 2304–2308 (1990)
Holzman, T.F., Egan, D.A., Edalji, R., Simmer, R.L., Helfrich, R., Taylor, A., Burres, N.S.: Preliminary characterization of a cloned neutral isoelectric form of the human peptidyl prolyl isomerase cyclophilin. J. Biol. Chem. 266, 2474–2479 (1991)
Wang, M.Z., Shetty, J.T., Howard, B.A., Campa, M.J., Patz, E.F., Fitzgerald, M.C.: Thermodynamic analysis of Cyclosporin A binding to Cyclophilin A in a lung tumor tissue lysate. Anal. Chem. 76, 4343–4348 (2004)
Liu, J., Albers, M.W., Wandless, T.J., Luan, S., Alberg, D.G., Belshaw, P.J., Cohen, P., Mackintosh, C., Klee, C.B., Schreiber, S.L.: Inhibition of T cell signaling by imunophilin-ligand complexes correlates with loss of calcineurin phosphatase activity. Biochemistry 31, 3896–3901 (1992)
Liu, J., Farmer Jr., J.D., Lane, W.S., Friedman, J., Weissman, I., Schreiber, S.L.: Calcineurin is a common target of cyclophilin-cyclosporin A and Fkbp-Fk506 complexes. Cell 66, 807–815 (1991)
Jin, L., Harrison, S.C.: Crystal structure of human calcineurin complexed with cyclosporin A and human cyclophilin. Proc. Natl. Acad. Sci. U. S. A. 99, 13522–13526 (2002)
Acknowledgments
The authors acknowledge support for this work by a grant from the National Institutes of Health (GM084174) to M.C.F.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
DeArmond, P.D., West, G.M., Huang, HT. et al. Stable Isotope Labeling Strategy for Protein–Ligand Binding Analysis in Multi-Component Protein Mixtures. J. Am. Soc. Mass Spectrom. 22, 418–430 (2011). https://doi.org/10.1007/s13361-010-0060-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-010-0060-1