Abstract
The application of the direct electrospray ionization mass spectrometry (ESI-MS) assay to quantify interactions between bovine β-lactoglobulin (Lg) and a series of fatty acids (FA), CH3(CH2)xCOOH, where x = 6 (caprylic acid, CpA), 8 (capric acid, CA), 10 (lauric acid, LA), 12 (myristic acid, MA), 14 (palmitic acid, PA) and 16 (stearic acid, SA), is described. Control ESI-MS binding measurements performed on the Lg-PA interaction revealed that both the protonated and deprotonated gas phase ions of the (Lg + PA) complex are prone to dissociate in the ion source, which leads to artificially small association constants (K a ). The addition of imidazole, a stabilizing solution additive, at high concentration (10 mM) increased the relative abundance of (Lg + PA) complex measured by ESI-MS in both positive and negative ion modes. The K a value measured in negative ion mode and using sampling conditions that minimize in-source dissociation is in good agreement with a value determined using a competitive fluorescence assay. The K a values measured by ESI-MS for the Lg interactions with MA and SA are also consistent with values expected based on the fluorescence measurements. However, the K a values measured using optimal sampling conditions in positive ion mode are significantly lower than those measured in negative ion mode for all of the FAs investigated. It is concluded that the protonated gaseous ions of the (Lg + FA) complexes are kinetically less stable than the deprotonated ions. In-source dissociation was significant for the complexes of Lg with the shorter FAs (CpA, CA, and LA) in both modes and, in the case of CpA, no binding could be detected by ESI-MS. The affinities of Lg for CpA, CA, and LA determined using the reference ligand ESI-MS assay, a method for quantifying labile protein–ligand complexes that are prone to in-source dissociation, were found to be in good agreement with reported values.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
The direct electrospray ionization mass spectrometry (ESI-MS) assay has emerged as a useful technique for measuring the binding stoichiometry and affinity (K a ) of protein–ligand interactions, as well as other noncovalent biological complexes, in solution. The assay is based on the relative abundance of free and ligand-bound protein ions measured by ESI-MS for a solution of known initial protein and ligand concentration [1–3]. The ESI-MS assay is fast (measurements normally can be completed in less than ~1 min), consumes little sample (typically ~ pmol per analysis when performed using nanoflow ESI), provides direct insight into binding stoichiometry and can be used to measure multiple binding equilibria simultaneously. Furthermore, the assay is very versatile as there is no requirement for labeling or immobilization of analyte.
Absolute K a values, in the 103–107 M−1 range, have been determined for a variety of protein–ligand complexes, including antibody-antigen, lectin-carbohydrate, enzyme-inhibitor complexes, and shown to be in reasonable agreement with values determined by other analytical methods [4–10]. To date, however, there have been few reported examples of the application of ESI-MS to characterize protein-hydrophobic ligand binding [11–14] and, to our knowledge, no reports of absolute affinity measurements using the direct ESI-MS assay. The two principal challenges to quantifying interactions between water soluble proteins and hydrophobic ligands using ESI-MS are the low solubility of the ligand in aqueous solution, and the propensity of the desolvated complexes to dissociate during analysis. Low ligand solubility prevents the ESI-MS assay from being applied to very weak interactions (<104 M−1) because high ligand concentrations are needed to produce detectable levels of complex. Protein-ligand complexes stabilized predominantly by nonpolar interactions also tend to have low kinetic stabilities in the gas phase and are prone to dissociate in the ion source (i.e., in-source dissociation) [11, 12]. In-source dissociation reduces the relative abundance of the ligand-bound protein ions and, consequently, leads to artificially low binding constants [11, 15–17]. In extreme cases, in-source dissociation results in false negatives, wherein no ligand-bound protein ions are detected by ESI-MS [15]. However, using gentle sampling conditions, along with stabilizing solution or gas phase additives, it is sometimes possible to preserve the labile protein–ligand complexes during ESI-MS analysis and to quantify their association constants [17, 18].
Here, we describe the application of ESI-MS to quantify the affinities of bovine β-lactoglobulin (Lg) for a series of linear, saturated fatty acids (FA), CH3(CH2)xCOOH. Lg, a member of the lipocalin protein family, possesses a large hydrophobic cavity and is known to bind a variety of hydrophobic ligands, including FAs [19, 20]. Association constants for the interaction between Lg and several FAs [x = 6 (caprylic acid), 10 (lauric acid), and 14 (palmitic acid)], at 25 °C and various pH values (6.2, 7.2, and 8.3) have been determined using a competitive fluorescence assay [21]. Notably, these measurements revealed that at a given pH, the logarithm of K a increases linearly with the length of the FA acyl chain. Based on this dependence, it is possible to estimate the K a values for related FAs, vide infra. To test the reliability of the direct ESI-MS assay for quantifying protein–hydrophobic ligand interactions, affinity measurements were carried out on Lg and a series of FAs (x = 6, 8, 10, 12, 14, and 16) at pH 8.5 and 25 °C. Importantly, it is shown that the K a values for Lg interactions with the longer FAs (x = 12, 14, and 16) measured in negative ion mode and using gentle sampling conditions in conjunction with a stabilizing solution additive are in good agreement with the reported values. In contrast, in-source dissociation of the gaseous ions of the complexes of Lg and the shorter FAs is extensive, which precludes the determination of reliable affinities using the direct ESI-MS assay. However, the affinities determined for the shorter FAs using the reference ligand ESI-MS assay [21], a method for quantifying labile protein–ligand complexes that are prone to in-source dissociation, are in good agreement with reported values.
2 Experimental
2.1 Protein and Ligand
Bovine β-lactoglobulin (Lg, monomer MW 18281 Da), caprylic acid (CpA, 144.2 Da), capric acid (CA, 172.3 Da), lauric acid (LA, 200.3 Da), myristic acid (MA 228.4 Da), palmitic acid (PA, MW 256.4 Da), and stearic acid (SA, 284.8 Da) were purchased from Sigma-Aldrich Canada (Oakville, Canada). The Lg was dissolved and exchanged directly into Milli-Q water, using an Amicon microconcentrator with a molecular weight cutoff of 10 kDa, followed by filtration using an Amicon ultra centrifugal filter with a molecular weight cutoff of 100 kDa to remove Lg aggregates in solution. The concentration of the Lg solution was determined by lyophilizing a known volume of the filtrate and measuring the mass of the protein. The protein stock solution was stored at −20 °C. The ligand stock solutions were prepared by dissolving each FA into aqueous ammonium acetate (25 mM). The ESI solutions were prepared from aqueous stock solutions of protein and ligand. Aqueous ammonium hydroxide was added to adjust the pH of the ESI solution to 8.5. Where indicated, imidazole (10 mM) was also added in order to stabilize the (Lg + FA) complexes during ESI-MS analysis.
2.2 Mass Spectrometry
All experiments were performed on a 9.4 tesla Apex II Fourier transform ion cyclotron resonance (FTICR) mass spectrometer (Bruker, Billerica, MA) equipped with a nanoflow ESI ion source. NanoES was performed using borosilicate tubes (1.0 mm o.d., 0.68 mm i.d.), pulled to ~5 μm o.d. at one end using a P-2000 micropipette puller (Sutter Instruments, Novato, CA). Details of the instrumental parameters employed in positive ion mode are given below. For measurements in negative ion mode, the voltage polarity was switched. The electric field required to spray the solution in positive ion mode was established by applying a voltage of 800 V to a platinum wire inserted inside the glass tip. The solution flow rate was ~20 nL/min. The gaseous ions produced by nanoES were introduced into the mass spectrometer through a stainless steel capillary (i.d. 0.43 mm) maintained at an external temperature of 66 °C. The gas flow rate into the instrument was measured to be 0.6 L min−1. The ions sampled by the capillary (50 V) were transmitted through a skimmer (0 V) and accumulated electrodynamically in an rf hexapole. Ions were then ejected from the hexapole and accelerated (−2700 V) into the superconducting magnet, decelerated, and introduced into the ion cell. The trapping plates of the cell were maintained at a constant potential of 1.4 V throughout the experiment. The typical base pressure for the instrument was ~5 × 10−10 mbar. Data acquisition was controlled by an SGI R5000 computer running the Bruker Daltonics
XMASS software, ver. 5.0. Mass spectra were obtained using standard experimental sequences with chirp broadband excitation. The time domain signal, consisting of the sum of 50 transients containing 128 K data points per transient, was subjected to one zero-fill prior to Fourier transformation.
2.3 Determining Protein–Ligand Affinities Using the Direct ESI-MS Assay
The procedures used for the determination of protein–ligand binding constants (K a ) directly from ES mass spectra are described in detail elsewhere [4]. Briefly, the direct ESI-MS assay is based on the quantification of the abundance (Ab) of ligand-bound and unbound protein ions in the gas phase, e.g., PLn+ and Pn+, respectively. The measured abundance ratio (R) is assumed to be equivalent to the equilibrium concentration ratio of ligand-bound and free protein in solution, Eq. 1:
From the measured R value and initial concentrations of protein ([P]o) and ligand ([L]o), K a can be calculated using Eq. 2:
2.4 Determining Protein–Ligand Affinities Using the Reference Ligand ESI-MS Method
The reference ligand ESI-MS method for determining protein–ligand affinities is based on the direct ESI-MS assay and competitive ligand binding [22]. A reference ligand (L ref ), which binds specifically to the protein, at the same binding site as the ligand of interest, with known affinity and forms a stable protein-ligand complex in the gas phase, is added to the solution. The fraction of P bound to L ref , which is determined directly from the ESI mass spectrum, is sensitive to the fraction of P bound to L in solution and enables the affinity of P for L to be determined.
In the absence of in-source dissociation, the measured abundance ratios of ligand-bound to free P ions (R and R ref ) will reflect the concentration ratios in solution and K a for the PL complex can be calculated using Eq. 3:
where R is given by Eq. 1 and R ref by Eq. 4:
If the gas phase ions of the PL complex are kinetically labile and undergo in-source dissociation, the relative abundance of ligand-bound and free protein ions measured by ESI-MS will no longer reflect the original concentration ratios. Instead, the measured abundance ratio of L ref -bound P to free P gas phase ions (i.e., R ref,app ) will depend on the concentration of both free P and PL originally present in solution, Eq. 5:
where α is the fraction of PL that undergoes in-source dissociation.
In the case of complete dissociation of PL, i.e., α = 1, R ref (the “true” concentration ratio [PLref]/[P]) can be calculated from R ref,app and Ka,ref using Eq. 6:
and R can be calculated with Eq. 7:
Once R and R ref are known, the K a value for the PL complex can be calculated using Eq. 3.
If the gas phase PL ions undergo partial dissociation in the source, i.e., α < 1, the measured abundance ratio of L-bound P to free P gas phase ions (i.e., R app ) can be determined using Eq. 8:
Therefore, R ref can be determined from the measured R ref,app and R app values using Eq. 9:
and R can be found using Eq. 10:
Once R and R ref are known, K a value for the PL complex can be calculated using Eq. 3.
3 Results and Discussion
3.1 Association Constant for the (Lg + PA) Complex Determined by the Direct ESI-MS Assay
The (Lg + PA) complex served as a model protein-fatty acid complex for establishing appropriate experimental and instrumental conditions for the direct ESI-MS binding measurements. Shown in Figure 1a is an ESI mass spectrum acquired in positive ion mode for an aqueous solution of Lg (12 μM) and PA (10 μM) at pH 8.5 and 25 °C. At this pH and temperature, Lg is present only as monomer in solution [23]. Inspection of the mass spectrum reveals abundant signal corresponding to protonated Lgn+ ions, at n = 7 and 8, and protonated 1:1 complex, (Lg + PA)n+, at the same charge states. Notably, the K a value, 1.2 × 105 M−1, which was calculated from the measured abundance ratio (R) of ligand-bound to unbound Lg ions, is roughly 5-fold smaller than the reported affinity [21]. As described previously, collisional heating of gaseous ions can occur during accumulation in the rf hexapole in the FTICR mass spectrometer used for the present measurements [17]. Indeed, it was found that R (and K a ) increased as the accumulation time decreased. For example, the mass spectrum shown in Figure 1a was acquired using an accumulation time of 0.8 s, while the spectrum in Figure 1b was acquired for the same solution but using an accumulation time of 0.4 s. The decrease in accumulation time led to an increase of 47% in the magnitude of R, which corresponds to an increase of 83% in the K a value, 2.2 × 105 M−1. The effect of accumulation time on the binding measurements is more clearly seen in Figure 2, where the average R values are plotted versus accumulation time (from 0.4 to 2.0 s). It can be seen that the magnitude of R decreases systematically with increasing accumulation time and is immeasurably small at times ≥2.0 s. These results indicate that, even using optimal instrumental conditions and the shortest accumulation time (0.4 s) that still provides sufficient protein ion signal, the relative abundance of the (Lg + PA)n+ ions measured by ESI-MS is affected (reduced) by in-source dissociation.
ES mass spectra obtained in positive ion mode for aqueous ammonium acetate (10 mM, pH 8.5) solutions of (a) Lg (12 μM), PA (10 μM), accumulation time 0.8 s; (b) Lg (12 μM), PA (10 μM), 0.4 s; (c) Lg (12 μM), PA (10 μM), imidazole (10 mM), 0.8 s; (d) Lg (12 μM), PA (10 μM), imidazole (10 mM), 0.4 s, and in negative ion mode for aqueous ammonium acetate (10 mM, pH 8.5) solutions of (e) Lg (12 μM), PA (10 μM), imidazole (10 mM), accumulation time 0.8 s, and (f) Lg (12 μM), PA (10 μM), imidazole (10 mM), 0.4 s. The solution temperature for all measurements was 25 °C
Influence of hexapole accumulation time on the R value determined by ESI-MS in positive ion mode for aqueous ammonium acetate (10 mM, pH 8.5) solutions of Lg (12 μM) and PA (10 μM), (■); Lg (12 μM), PA (10 μM) and imidazole (10 mM), (▲); and in negative ion mode for an aqueous ammonium acetate (10 mM, pH 8.5) solution of Lg (12 μM), PA (10 μM), imidazole (10 mM), (●). The dashed line indicates the expected R value based on the Ka value reported in reference [21]. The solution temperature for all measurements was 25 °C
Recently, it was shown that the presence of imidazole, at high concentration (~10 mM), can protect protein-ligand complexes from in-source dissociation during ESI-MS analysis [17]. It was proposed that the stabilizing effect is due, at least in part, to enhanced evaporative cooling, which arises from the loss of multiple nonspecifically bound imidazole molecules from the gaseous protein–ligand complexes in the ion source [17]. The stabilizing effect of imidazole is evident in the ESI mass spectra shown in Figure 1c and d. These mass spectra were acquired under identical conditions as those shown in Figure 1a and b, respectively, but with the addition of 10 mM imidazole to the solution. The presence of imidazole results in two obvious changes in the appearance of the mass spectra. The charge state distribution for the Lgn+ and (Lg + PA)n+ ions broadens from 7–8 to 6–8. This change is due to the high gas phase basicity of imidazole (217 kcal mol−1) [24] and its ability to strip protons from multiply protonated protein ions in the gas phase [25]. Additionally, the relative abundance of the (Lg + PA)n+ ions is larger than that measured in the absence of imidazole. In fact, the R values measured at 0.4 and 0.8 s hexapole accumulation times are nearly indistinguishable. However, the corresponding K a value, (2.7 ± 0.2) × 105 M−1, is still a factor of two smaller than the reported value, suggesting the possibility that in-source dissociation was not completely eliminated under these conditions.
The ESI-MS binding measurements were also carried out in negative ion mode. Shown in Figure 1e and f are mass spectra acquired in negative ion mode for a solution of Lg (12 μM) and PA (10 μM) using accumulation times of 0.8 and 0.4 s, respectively. Imidazole (10 mM) was also added to minimize the occurrence of in-source dissociation. Inspection of the mass spectra reveals signal corresponding to deprotonated Lgn– and (Lg + PA)n– ions, at n = 6 to 8. Notably, the corresponding R values are significantly larger than those measured in positive ion mode under the same conditions (Figure 2). Furthermore, the average K a value (3.8 ± 0.1) × 105 M−1, which was obtained from replicate measurements performed at four different concentrations, is in good agreement with the reported value of (5.0 ± 0.2) × 105 M−1 (Table 1) [21].
The larger R values measured in negative ion mode compared with those measured in positive ion mode confirm that, even with the addition of imidazole to the solution, a fraction of the protonated (Lg + PA)n+ ions are lost due to in-source dissociation. At present, the origin of the differential stabilities of the protonated and deprotonated ions of the (Lg + PA) complex is not known but presumably reflects differences in the structure of the Lg ions arising from differences in the nature of the charges and their locations. Our laboratory is currently carrying out a comparative study, employing thermal dissociation rate constant measurements and molecular dynamics simulations, of the structures and stabilities of protonated and deprotonated (Lg + FA) ions. The results of this study will be reported in a forthcoming manuscript.
3.2 Association Constants for the (Lg + FA) Complexes Determined by the Direct ESI-MS Assay
Having established optimal experimental/instrumental conditions for quantifying the interaction between Lg and PA, the direct ESI-MS assay was used to measure binding constants for the interactions between Lg and the other FAs (CpA, CA, LA, MA, and SA). Shown in Figure 3 are illustrative ES mass spectra obtained in negative ion mode for aqueous solutions (pH 8.5) of Lg with one of the five other FAs, at ligand concentration ranging from 10 to 52 μM. Again, imidazole (10 mM) was added to the solutions to protect against in-source dissociation. Ion signal corresponding to the deprotonated (Lg + FA) complexes was detected for all of the FAs except for CpA, where no ions corresponding to the (Lg + CpA) complex were observed. Increasing the concentration of CpA from 52 to 100 μM (where 70% of Lg is expected to be bound to CpA, based on the reported affinity of 2.6 × 104 M−1) [21] gave similar results. The complete absence of signal corresponding to the (Lg + CpA) complex indicates that the gaseous ions are highly susceptible to in-source dissociation.
ESI mass spectra acquired for aqueous ammonium acetate (10 mM, pH 8.5) solutions of (a) Lg (12 μM), CpA (52 μM), (b) Lg (12 μM), CA (51 μM), (c) Lg (10 μM), LA (30 μM), (d) Lg (10 μM), MA (22 μM), and (e) Lg (11 μM), SA (10 μM). Each solution contained 10 mM imidazole. The solution temperature for all measurements was 25 °C
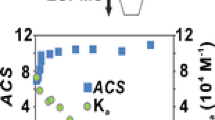
The average K a values determined for CA, LA, MA, and SA, from replicate measurements performed at multiple ligand concentrations, are listed in Table 1. In order to more easily compare the K a values measured by ESI-MS with the values expected based on fluorescence measurements, the logarithm of K a (determined by either ESI-MS or competitive fluorescence) is plotted versus FA acyl chain length in Figure 4. It can be seen that the K a values determined by ESI-MS for the longer FAs, MA, and SA, are in excellent agreement with the values expected based on the linear trend established from the fluorescence-derived K a values [21]. In contrast, the K a value measured for LA is only 25% of the reported value; the deviation is even more pronounced for CA, where the K a is only 8% of the reported value. The discrepancy in the K a values indicates that the deprotonated ions of (Lg + CA) and (Lg + LA) undergo in-source dissociation. The susceptibility of the deprotonated ions of the (Lg + FA) complexes composed of the shorter FAs to in-source dissociation can be explained, at least in part, by the results of recent time-resolved thermal dissociation experiments performed on deprotonated (Lg + FA)7– ions composed of LA, MA, PA and SA [26]. In this study, it was found that the gaseous (Lg + FA)7– ions adopt one of two kinetically distinct structures, referred to as the fast and slow components. In the fast component, the FA ligand is stabilized predominantly by protein–lipid interactions, while for slow component, H-bonds between the ligand carboxyl group and Lg also contribute to the stability of the complex. Furthermore, over the temperature range investigated (26 to 66 °C), the kinetic stabilities of the fast components decreased with decreasing acyl chain length [26]. Based on these results, it is reasonable to expect that the effect of in-source dissociation on the relative abundance of the (Lg + FA)n– ions will increase with decreasing acyl chain length.
Plot of logarithm of K a measured by direct ESI-MS assay (▲), reference ligand ESI-MS method (Δ) and competitive fluorescence assay (●) versus number of methylene groups (x) for the (Lg + CH3(CH2)xCOOH) complexes. The dashed line corresponds to linear least squares fit of the log K a values measured by fluorescence [21]. The ESI-MS binding measurements were performed at pH 8.5 and 25 °C; the fluorescence data were determined at pH 8.3 and 25 °C
3.3 Association Constants for the (Lg + FA) Complexes Determined by the Reference Ligand ESI-MS Assay
Recently, our laboratory developed a competitive ligand binding assay for the quantification of protein–ligand interactions that are labile in the gas phase and undergo partial or complete in-source dissociation during ESI-MS analysis [22]. The method, referred to as the reference ligand ESI-MS assay, was used in the present study to quantify the interactions between Lg and CpA, CA, and LA. To implement the assay, an appropriate L ref is needed. As discussed above, the similarity in the binding constants measured for the (Lg + PA) complex directly by ESI-MS in negative ion mode and by fluorescence suggests that in-source dissociation of the (Lg + PA)n– ions was negligible under the experimental conditions used. Therefore, PA was deemed a suitable L ref for these measurements. Shown in Figure 5a is an ESI mass spectrum acquired for a solution of Lg (12 μM), PA (10 μM), and imidazole (10 mM); the mass spectrum shown in Figure 5b was obtained for the same solution as above but with the addition of CpA (52 μM). Although ions corresponding to the (Lg + CpA) complex were not detected, the addition of CpA to the solution resulted in a decrease in the fraction of Lg bound to PA, indicating the presence of specific binding between Lg and CpA in solution. Plotted in Figure 6 is the fraction of bound (to PA) and unbound Lg determined by direct ESI-MS measurements performed at fixed concentrations of Lg (12 μM) and PA (10 μM) and varying concentrations of CpA (0, 39, 52, and 65 μM). Notably, the fraction of Lg bound to PA decreased with increasing concentration of CpA. Analysis of the ESI-MS data using the approach described above yields an average K a value of (2.3 ± 0.3) × 104 M−1 for the (Lg + CpA) complex (Table 1). Importantly, this value is indistinguishable, within experimental error, from the fluorescence-derived value of (2.6 ± 0.5) × 104 M−1 [21].
Distribution of the relative abundance of Lg (≡ P) and (Lg + PA) complex (≡ PL ref ) measured by ESI-MS in negative ion mode for aqueous ammonium acetate (10 mM, pH 8.5) solutions of Lg (12 μM), PA (10 μM), and CpA at concentrations of 0, 39, 52, and 65 μM. Each solution contained 10 mM imidazole. The solution temperature for all measurements was 25 °C
The reference ligand ESI-MS method was also used to establish the binding constants for the (Lg + CA) and (Lg + LA) complexes. Again, PA was used as L ref for these measurements. Shown in Figure S1, which can be found in the electronic version of this article, are illustrative ES mass spectra acquired for solutions of Lg and PA, in the absence and presence of either CA or LA. As outlined above, data analysis was carried out differently than was the case for the (Lg + CpA) complex because ions corresponding to the (Lg + CA) and (Lg + LA) complexes could still be detected [22]. The binding constant determined for (Lg + LA) complex using the reference ligand ESI-MS assay, (1.1 ± 0.2) × 105 M−1, is in excellent agreement with the reported value, (1.3 ± 0.1) × 105 M−1. The value measured for (Lg + CA) complex, (4.6 ± 0.5) × 104 M−1, is also in reasonable agreement with the value (6 × 104 M−1) estimated from the linear dependence of log K a on acyl chain length (Figure 4).
4 Conclusions
In summary, the use of ESI-MS to quantify protein–fatty acid complexes in aqueous solution is described. The interactions between Lg and a series of linear, saturated FAs (CpA, CA, LA, MA, PA, and SA) were investigated using both the direct ESI-MS assay and the reference ligand ESI-MS method. The binding constant measured for PA at 25 °C and pH 8.5 using the direct ESI-MS assay, performed in negative ion mode and utilizing a stabilizing solution additive (imidazole), is in good agreement with the value measured using a competitive fluorescence assay. The affinities measured directly for MA and SA are also consistent with values expected based on the linear dependence of the log K a values with acyl chain length established from the fluorescence-derived binding data for related FAs. However, the K a values measured directly for LA and CA were significantly smaller than the expected values. In the case of CpA, no binding was detected by direct ESI-MS measurement. These results indicate the occurrence of in-source dissociation, which reduces the relative abundance of the gaseous (Lg + FA)n- ions, for these shorter FAs. Notably, in-source dissociation was prevalent in the ESI-MS binding measurements performed in positive ion mode for all of the (Lg + FA) complexes investigated. The reference ligand ESI-MS assay was used to measure binding constants for the Lg interactions with the shorter FAs. Importantly, the measured K a values for CpA and LA are in excellent agreement with values measured using the fluorescence assay.
References
Daniel, J.M., Friess, S.D., Rajagopalan, S., Wendt, S., Zenobi, R.: Quantitative determination of noncovalent binding interactions using soft ionization mass spectrometry. Int. J. Mass Spectrom. 216, 1–27 (2002)
Schug, K.A.: Solution phase enantioselective recognition and discrimination by electrospray ionization-mass spectrometry: State-of-the-Art methods and an eye towards increased throughput measurements. Comb. Chem. High Throughput Screen. 10, 301–316 (2007)
Heck, A.J.R., Van Den Heuvel, R.H.H.: Investigation of intact protein complexes by mass spectrometry. Mass Spectrom. Rev. 23, 368–389 (2004)
Wang, W., Kitova, E.N., Klassen, J.S.: Influence of solution and gas phase processes on protein-carbohydrate binding affinities determined by nanoelectrospray Fourier-transform ion cyclotron resonance mass spectrometry. Anal. Chem. 75, 4945–4955 (2003)
Shoemaker, G.K., Soya, N., Palcic, M.M., Klassen, J.S.: Temperature-dependent cooperativity in donor-accepator substrate binding to the human blood group glycosltransferases. Glycobiology 18, 587–592 (2008)
Soya, N., Shoemaker, G.K., Palcic, M.M., Klassen, J.S.: Comparative study of substrate and product binding to the human ABO(H) blood group glycosyltransferases. Glycobiology 19, 1224–1234 (2009)
Rademacher, C., Shoemaker, G.K., Kim, H.-S., Zheng, R.B., Taha, H., Liu, C., Nacario, R.C., Schriemer, D.C., Klassen, J.S., Peters, T., Lowary, T.L.: Ligand specificity of CS-35, a monoclonal antibody that recognizes mycobacterial lipoarabinomannan. A model system for oligofuranoside-protein recognition. J. Am. Chem. Soc. 129, 10489–10502 (2007)
Jorgensen, T.J.D., Roepstorff, P., Heck, A.J.R.: Direct determination of solution binding constants for noncovalent complexes between bacterial cell wall peptide analogues and vancomycin group antibiotics by electrospray ionization mass spectrometry. Anal. Chem. 70, 4427–4432 (1998)
Jecklin, M.C., Touboul, D., Bovet, C., Wortmann, A., Zenobi, R.: Which electrospray-based ionization method best reflects protein-ligand interactions found in solution? A comparison of ESI, nanoESI, and ESSI for the determination of dissociation constants with mass spectrometry. J. Am. Soc. Mass Spectrom. 19, 332–343 (2008)
Yu, Y.H., Kirkup, C.E., Pi, N., Leary, J.A.: Characterization of noncovalent protein-ligand complexes and associated enzyme intermediates of GlcNAc-6-O-sulfotransferase by electrospray ionization FTICR mass spectrometry. J. Am. Soc. Mass Spectrom. 15, 1400–1407 (2004)
Robinson, C.V., Chung, E.W., Kragelund, B.B., Knudsen, J., Aplin, R.T., Poulsen, F.M., Dobson, C.M.: Probing the nature of noncovalent interactions by mass spectrometry. A study of protein-CoA ligand binding and assembly. J. Am. Chem. Soc. 118, 8646–8653 (1996)
Yin, S., Xie, Y., Loo, J.A.: Mass spectrometry of protein–ligand complexes: Enhanced gas-phase stability of ribonuclease-nucleotide complexes. J. Am. Soc. Mass Spectrom. 19, 1199–1208 (2008)
Stanley, W.A., Versluis, K., Schultz, C., Heck, A.J.R., Wilmanns, M.: Investigation of the ligand spectrum of human sterol carrier protein 2 using a direct mass spectrometry assay. Arch. Biochem. Biophys. 461, 50–58 (2007)
Hooper, A.M.; Dufour, S.; He, X.; Muck, A.; Zhou, J.; Almeida, R.; Field, L.M.; Svatos, A.; Pickett, J.A. High-throughput ESI-MS analysis of binding between the Bombyx mori pheromone-binding protein BmorPBP1, its pheromone components, and some analogues. Chem. Commun. 5725–5727 (2009)
Clark, S.M., Konermann, L.: Determination of ligand−protein dissociation constants by electrospray mass spectrometry-based diffusion measurements. Anal. Chem. 76, 7077–7083 (2004)
Van Dongen, W.D., Heck, A.J.R.: Binding of selected carbohydrates to apo-concanavalin a studied by electrospray ionization mass spectrometry. Analyst 125, 583–589 (2000)
Sun, J., Kitova, E.N., Klassen, J.S.: Method for stabilizing protein–ligand complexes in nanoelectrospray ionization mass spectrometry. Anal. Chem. 79, 416–425 (2007)
Bagal, D., Kitova, E.N., Liu, L., El-Haweit, A., Schnier, P.D., Klassen, J.S.: Gas phase stabilization of noncovalent protein complexes formed by electrospray ionization. Anal. Chem. 81, 7801–7806 (2009)
Kontopidis, G., Holt, C., Sawyer, L.: Beta-lactoglobulin: Binding properties, structure, and function. J. Dairy Sci. 87, 785–796 (2004)
Qin, B.Y., Bewley, M.C., Creamer, L.K., Baker, H.M., Baker, E.N., Jameson, G.B.: Structural basis of the tanford transition of bovine β-lactoglobulin. Biochemistry 37, 14014–14023 (1998)
Collini, M., D’Alfonso, L., Molinari, H., Ragona, L., Catalaono, M., Baldini, G.: Competitive binding of fatty acids and the fluorescent probe 1-8-anilinonaphthalene sulfonate to bovine β-lactoglobulin. Protein Sci. 12, 1596–1603 (2003)
El-Hawiet, A., Kitova, E.N., Liu, L., Klassen, J.S.: Quantifying labile protein–ligand interactions using electrospray ionization mass spectrometry. J. Am. Soc. Mass Spectrom. 21, 1893–1899 (2010)
Wu, S.-Y., Perez, M.D., Puyol, P., Sawyer, L.: β-lactoglobulin binds palmitate within its central cavity. J. Biol. Chem. 274, 170–174 (1999)
Hunter, E.P., Lias, S.G.: Evaluated gas phase basicities and proton affinities of molecules: An update. J. Phys. Chem. Ref. Data 27, 413–656 (1998)
Kitova, E.N., Seo, M., Roy, P.N., Klassen, J.S.: Elucidating the intermolecular interactions within a desolvated protein-ligand complex. An experimental and computational study. J. Am. Chem. Soc. 130, 1214–1226 (2008)
Liu, L., Bagal, D., Kitova, E.N., Schnier, P.D., Klassen, J.S.: Hydrophobic protein–ligand interactions preserved in the gas phase. J. Am. Chem. Soc. 131, 15980–15981 (2009)
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOC 168 kb)
Rights and permissions
About this article
Cite this article
Liu, L., Kitova, E.N. & Klassen, J.S. Quantifying Protein-Fatty Acid Interactions Using Electrospray Ionization Mass Spectrometry. J. Am. Soc. Mass Spectrom. 22, 310–318 (2011). https://doi.org/10.1007/s13361-010-0032-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13361-010-0032-5