Abstract
Role of efflux-mediated toxin resistance to trichothecenes is known in trichothecene-producing species. However, the role of trichothecene efflux pump homologues in non-producing fusaria such as F. oxysporum and F. proliferatum was not investigated in detail. Analysis of the homologues of trichothecene efflux pump from multiple fungal species allowed us to uncover and catalogue functional gene copies of conserved structure. Putative Tri12 candidates in Fusarium oxysporum and F. proliferatum were characterised via expression profiling in response to different trigger compounds, providing supporting evidence for role of Tri12 homologues in the resistance to trichothecenes. Our analysis of Tri12 phylogeny also suggests that efflux-mediated trichothecene resistance is likely to predate the divergence of Trichoderma and Fusarium species. On the regulatory level, we posit that the increased tolerance of trichothecenes by F. oxysporum is possibly related to the decoupling of Tri12 homologue expression from pH, due to the deletion of PACC/RIM101 transcription factor binding site in its promoter region.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mycotoxins are bioactive fungal secondary metabolites which are typically viewed mainly through the lens of their harmful properties to humans and livestock. Nevertheless, a large array of fungal pathogens spend most of their life cycle, in competition with prokaryotic and eukaryotic microbes (either persisting in the soil environment or competing for plant hosts). Thus, one can expect that many compounds affect fungal fitness through their effect on the microbe-microbe competition. In cases of some toxins, the divergence and spread of different toxigenic properties have been shown to predate the origins of major classes of filamentous fungi (e.g. Koczyk et al. 2015), with detoxification mechanisms likewise being ancient (e.g. Popiel et al. 2014; Perlin et al. 2014). In the different environments, the competitive impact of toxin biosynthesis can be positive, due to increased isolate virulence or toxin’s inhibitory effects on the growth of other strains (Losada et al. 2009).
In this context, there is evidence demonstrating that trichothecene biosynthesis has impact on fungus-fungus competition. For example, Ramakrishna et al. (1996) found that during competition between F. sporotrichioides (producer of T-2 toxin) and two other fungi: A. flavus and Penicillium verrucosum the growth of F. sporotrichioides was negatively affected but paradoxically the production of T-2 mycotoxin was stimulated. Similarly, Lutz et al. (2003) tested the impact of deoxynivalenol (DON) against a potent fungal antagonist Trichoderma atroviride and described decreased expression of genes coding chitinase in the presence of these toxins. In the studies of McLaughlin et al. (2009) and Bin-Umer et al. (2011), they observed the impact of trichothecenes on yeast cells. The results illustrated that mycotoxins can inhibit the mitochondrial membrane potential, translation and levels of reactive oxygen species in fungi, in a dose-dependent manner.
Trichothecene biosynthesis contributes to increased virulence of fungal strains, and further inquiries into other fungal species show that the basic trichothecene scaffold is likely ancient at least within the context of multiple species within the Hypocreales order (Trichoderma arundinaceum and Trichoderma brevicompactum—Cardoza et al. 2011; Myrothecium roridum—Trapp et al. 1998; Stachybotrys sp.—Semeiks et al. 2014). While capacity for trichothecene biosynthesis is present in multiple members of the Hypocreales order, the Fusarium genus is perhaps the best characterised group (Kimura et al. 2007). Fusarium spp. are subdivided into related, but phylogenetically distinct complexes that likely diverged in cretaceous period (O’Donnell et al. 2013). These fungi widely differ in their preferences in regards to saprophytic and/or pathogenic lifestyles as well as biosynthetic capabilities (trichothecenes are mainly produced by members of incarnatum-equiseti and sambucinum complexes). Frequently, the diverged species find themselves in direct or indirect competition when their ecological niches overlap. The relationship between F. graminearum and F. verticillioides on cereals (including variability in trichothecene accumulation) (Picot et al. 2012; Dawidziuk et al. 2016) provides one example of this phenomenon.
Mirroring the ancient origins of trichothecene biosynthesis, effective trichothecene resistance mechanisms are known to be partially present in multiple producing and non-producing strains (Kimura et al. 2003; Tokai et al. 2005; Proctor et al. 2009; Menke et al. 2012). A good example is trichotecene effux pump encoded by Tri12 geneor O-acetyltransferase Tri101 (and its divergent but functional Tri201 homologue). The Tri201 gene, in particular, was found to be present in both ancestrally divergent strains of Fusarium sp. from complexes other than sambucinum, e.g. the early diverging species F. decemcellulare, F. solani(Tokai et al. 2005) as well as in other species of fungi (Magnaporthe oryza—Tokai et al. 2005; Saccharomyces cerevisiae—Alexander et al. 2002).
While the trichothecene acetyltransferase homologues have been characterised in many species, the putative trichothecene efflux pump existence and functionality were not extensively investigated beyond the initial discovery of their functionality in the sambucinum complex (F. sporotrichioides—Alexander et al. 1999; F. graminearum—Wuchiyama et al. 2000). In producer species, the past comprehensive studies of Proctor and coworkers (Proctor et al. 2009; Cardoza et al. 2011) have shown that trichothecene efflux pump is frequently but not always present in the cluster (e.g. incarnatum-equiseti complex fusaria). More recently, a brief survey of Tri12 domain encoding transporters was conducted by Perlin et al. (2014), where homologues of unconfirmed function were summarised across many saprobic, animal and plant pathogenic-species.
To establish whether trichothecene efflux is a likely retained trait in previously not investigated species (F. oxysporum, F. proliferatum), we studied presence and evolutionary history of divergent Tri12 homologues. The research was performed by combining phylogenetic analyses of available Ascomycota sequences with gene expression and bioassays. Through the phylogenetic analysis of multiple Tri12 homologues, we confirmed the notion that active resistance to trichothecene-like compounds is likely an ancient trait or one common enough to elicit a convergent evolution of multiple, distantly related resistance factors (acetyltransferase and active transport). In order to obtain supporting evidence for functionality of divergent Tri12 homologues in fujikuroi and oxysporum complex, we investigated the expression of F. proliferatum and F. oxysporum homologues in response to varying stimuli (including trichothecene presence).
Methods
Isolate collection and identification
From the culture collections available at the Institute of Plant Genetics, Polish Academy of Sciences, Poznan, Poland, we selected the following strains lacking capacity for trichothecene biosynthesis: eight F. oxysporum strains (10 L, 11 L, 19 L, 55 L, 57 L, 94 L, 115 L, 131 L) with no recorded toxigenic potential (fumonisins, trichothecenes, zearalenone) and ten F. proliferatum strains (1 L, 3 L, 7 L, 21 L, 36 L, 58 L, 59 L, 66 L, 81 L, 99 L) known to produce fumonisins. Fungal strain annotation was conducted as per the protocol described in Dawidziuk et al. (2014) on basis of both morphological and molecular data: F. oxysporum strains 10 L—GenBank Accession number MN018756, 11 L-MN018757, 19 L-KF889103, 55 L-KF889104, 57 L-KF889105, 94 L-MN018758, 115 L-KF889099, 131 L-KF889101 and F. proliferatum strains 1 L-KF889131, 3 L-KF889134, 7 L-KF889137, 21 L-KF889122, 36 L-MN018759, 58 L-KF889136, 59 L-KF889132, 66 L-KF889125, 81 L-MN018760, 99 L-KF889127).
Bioassays with multiple compounds
For the purpose of the bioassay experiment, the concentration of deoxynivalenol was set to 8 mg/L as the lowest dose inhibiting fungal growth. The lower concentrations of toxin (1 mg/L, 2 mg/L and 5 mg/L) did not significantly influence the growth of the isolates and the higher doses (10 mg/L) inhibited growth of all fungal cultures.
The additional bioassay experiments were carried out to eliminate the impact of environmental factors on the growth of the tested cultures: MgCl2, KCl, ferulic acid, fungicide-Alert 350 SC (flusilazole) ground wheat seedlings, glucose, sucrose, coumaric acid, H2O2, caffeine, F. verticillioides (fumonisin producer). The concentration of additional chemical compounds was set to the same value as the concentration of DON. In the case of additional biological compounds, the 5 g of ground wheat was added to 250 mL of PDA medium and F. oxysporum/F. proliferatum bioassays with F. verticillioides was tested in the dual cultures (Gromadzka et al. 2009). The response was observed on PDA medium amended in simulated day (16 h)/night (8 h) conditions at 25 °C. The F. oxysporum assay was performed in three biological and ten technical replicates and the F. proliferatum assay was performed in three biological and ten technical replicates. Biological replicates were performed separately in the phytotron strictly controlling temperature, humidity and simulated day/night conditions.
The surface area of the fungal colonies was calculated by approximating the mycelium’s area to an ellipse by measuring both the length and width of the mycelium 4 days after toxin exposition (Dawidziuk et al. 2016).
Isolation of DNA and sequencing
Mycelium used for DNA extraction was obtained by inoculating Czapek-Dox broth (Sigma Aldrich, St. Louis, Missouri, USA) with yeast extract (Oxoid, Waltham, Massachusetts, USA) and streptomycin sulphate (50 mg/L, AppliChem, Darmstadt, Germany) and after incubation at 25 °C on a rotary shaker (120 rpm). Mycelium was collected on filter paper in a Büchner funnel, washed with sterile water, frozen at − 20 °C and freeze-dried. Total DNA was extracted using the DNeasy Plant Mini Kit (Qiagen, Hilden, Germany). The quality of DNA was estimated by a NanoDrop 2000 UV-Vis Spectrophotometer (Thermo Scientific, Wilmington, USA) and a Experion Automated Electrophoresis System (Bio-Rad, Hercules, CA). The protocols for primer design, PCR and sequencing conditions have been previously described by Popiel et al. (2014) and sequences of the primers are listed in Table 1.
Annotation of Tri12 homologues
The putative Tri12 homologues were gathered using a variant of the approach used in our previous work (Koczyk et al. 2015). Briefly, first a wide set of homologues was compiled through BLASTP searches against the combined NCBI/nr database (26/10/2015) and a local copy of Ensembl/Fungi release 28. The combined database was made non-redundant by clustering at 97% protein sequence identity with CD-HIT(Fu et al. 2012), representative sequences were inspected and kept on a per-species bases. The sequences of Tri12 from F. sporotrichioides and F. graminearum were used as queries.
To obtain the final set of Tri12 homologues, we performed unsupervised clustering with model transporters of known specificity. For this, the preliminary subset of candidates was combined with all available 2.A.1.3 (DHA14 antiporter family) homologues from Transporter Classification Data Base (Saier et al. 2016). The clustering was conducted in CLANS (Frickey and Lupas 2004) based on exhaustive all against all BLASTP comparisons with an expect value threshold of E-10. The stability of cluster containing Tri12 homologues was validated at more restrictive similarity thresholds (cluster membership was tested up to 1e-80 expect value threshold). The final set, used for alignment and phylogeny reconstruction, numbered a total of 33 sequences, after also including the two sequences corresponding to protein sequence consensi of, respectively, F. proliferatum and F. oxysporum Tri12 sequences obtained from the collection isolates.
We opted for the above iterative approach, as the simple selection, e.g. based on the conserved Pfam domain (TRI12) fingerprint, would result in a large set of poorly alignable distant homologues. Multiple sequences with Tri12 similarities are, upon inspection, DHA14 transporters of completely different specificity (such as STR1—the siderophore iron transporter from Schizosaccharomyces pombe, Pelletier et al. 2003) or present characteristic features of multidrug transporters (e.g. SGE1—violet/multidrug resistance protein from Saccharomyces cerevisiae, Ehrenhofer-Murray et al. 1998).
Where referenced, annotation of putative transcription factor binding sites was carried out in JASPAR (Mathelier et al. 2015). Putative transmembrane elements were annotated with CCTOP (Dobson et al. 2015) and TOPCONS (Tsirigos et al. 2015).
Sequence alignment and phylogeny reconstruction
The selected protein sequences were aligned with MAFFT-LINSI v 7.221 (Katoh and Standley 2013). For phylogenetic analysis, the multiple alignment was filtered with TCOFFEE/TCS module (Chang et al. 2014) using the transitive consistency score of 2 as the threshold (as recommended by the authors). The nucleotide sequences from the examined F. oxysporum (8 sequences) and F. proliferatum (10 sequences) isolates were aligned with MAFFT-LINSI and manually inspected for alignment correctness (referring to the earlier protein sequence alignment). For use in phylogeny reconstructions, a F. sporotrichioides reference Tri12 sequence as well as additional model F. oxysporum (4 sequences) and F. fujikuroi (1 sequence) sequences were added to this alignment.
Both nucleotide and protein, maximum likelihood phylogeny reconstructions were carried out with IQTREE v 1.3.6 (Nguyen et al. 2015), using built-in model selection and ultrafast bootstrap (Minh et al. 2013) procedure. In case of nucleotide sequences, this analysis was carried out in partitioned mode, with separate models for each exon and intron (auto-selected by IQTREE).
The full alignments of both nucleotide and protein sequences, used for phylogenetic reconstructions, are included in the Supplementary Materials to this article. The alignments were visualised in CLC Genomics Workbench v 8.5.1 (Qiagen) and the phylogenetic trees were drawn with MEGA (nucleotide sequence-based tree, Tamura et al. 2013) and ETE2 (protein tree, Huerta-Cepas et al. 2010). The relevant gene structures were annotated and visualised with WebScipio (Hatje et al. 2011).
Expression profiling
Mycelium was collected from the medium and each sample was weighed on a laboratory scale (due to rapid RNA degradation, wet weight was analysed) (Sartorius AG, Göttingen, Germany). Total RNA from chosen, representative isolates was purified using an RNeasy Plant Mini Kit (Qiagen, Hilden, Germany) according to the manufacturers’ protocol with the additional DNase digestion step. The quality of total RNA was estimated by Nanodrop (Thermo Scientific, Wilmington, DE) and via Bioanalyzer (Bio-Rad, Hercules, CA). RNA dissolved in DEPC water was stored at − 80 °C. qRT-PCR primers were designed on the basis of previously sequenced gene fragments using Primer 3 and their properties were tested using OligoCalc.
Real-time RT-PCR was used to amplify Tri12 homologues (trichothecene efflux pump) and Tri201 (homologue of trichothecene 3-O-acetyltransferse from F. graminearum) in F. oxysporum and F. proliferatum strains (Desjardins and Proctor 2007; Lee et al. 2011), and as a reference, we used housekeepeng genes Tub2 (β-tubulin), UBC(ubiquitin) and TEF1-α (translation elongation factor) from each RNA sample of the fungal strains.
Real-time RT-PCR reactions were performed using an CFX96 Touch™ Real-Time PCR Detection System (Bio-Rad, Hercules, CA). Analyses were conducted using iTaq One Step SYBR Green RT-qPCR Kit (Bio-Rad, Hercules, California, USA). The total reaction volume was 25 μL: 12.5 μL iTaq One Step SYBR Green RT-qPCR mix, 1 μL RNA (< 35 ng), 0.5 μL each primer (10 μM), 0.125 μL reverse transcriptase and 5.125 μL nuclease free water. The reaction was carried out using the following protocol: initial denaturation 94 °C for 2 min, followed by 40 cycles at 94 °C for 15 s, 59 °C for 1 min. In the experiment, we used three biological and two technical replicates together with a template-free negative control in each analysis of both target and control genes. The melting curve analysis (from 70 to 95 °C) confirmed primer pair specificity. As a control, we used mycelium samples cultivated on medium without the addition of toxins. Relative quantification of gene expression was calculated using the 2−ΔΔCt method (Bio-Rad, Hercules, CA). Data from samples treated with mycotoxin were normalised to β-tubulin, ubiquitin, 1-α translation elongation factor genes as internal controls (Real-Time PCR Application Guide, Bio-Rad, Hercules CA).
Statistical analyses
Statistical analyses of growth patterns (relative area on the Petri dish covered by fungus) comprised analyses of variance (ANOVA) and post hoc means comparisons (Tukey-Kramer honestly significant difference [HSD]; p ≤ 0.05) were performed with the Statistica 9.0 software package (Stat Soft, USA). The differences in gene expression between untreated and treated samples were analysed with Wilcoxon signed-rank non-parametric test (p ≤ 0.05) with use of one-tailed hypothesis. The test was performed on the delta Ct values from the second day after the exposition to the indicated compound(s).
Results
Growth patterns of strains treated with deoxynivalenol
Among all tested isolates of F. oxysporum and F. proliferatum, the response to deoxynivalenol in concentration of 8 mg/L was weak but still significant. Greater differences were noted between species. Growth of fumonisin producing F. proliferatum strains was inhibited by an average of 13% and of non-producingF. oxysporum by 6%. Importantly, in case of F. oxysporum isolates, one (10 L) was significantly different (more similar to F. proliferatum strains) and its growth was reduced by 10% (Table 2). Addition of lower doses of mycotoxin did not result in significant growth inhibition and higher concentration of deoxynivalenol in the medium suppressed the growth of all tested strains without any exceptions.
Growth of selected isolates in the control environment after addition of MgCl2, KCl, glucose, sucrose, F. verticillioides did not show significant changes (p ≤ 0.05) while the rest of the additives (ferulic acid, ground wheat seedlings, coumaric acid, H202, caffeine) caused significant reduction of the mycelium growth rate (Table 3).
Transcriptional response of Tri12 homologues in F. oxysporum and F. proliferatum
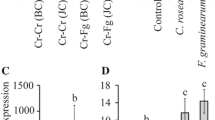
The expression of Tri12 homologue genes was analysed in F. oxysporum and F. proliferatum strains after deoxynivalenol treatment. As a reference, Tub2 (β-tubulin), UBC(ubiquitin) and TEF1-α (translation factor) genes were used. In the presence of mycotoxin, Tri12 gene in F. oxysporum strain (11 L) has shown significantly (P ≤ 0.05) increased transcriptional activity (13.45-fold change–96 h after toxin exposition). Weakest induction was observed in deoxynivalenol treated F. proliferatum isolate, in which the relative normalised expression of Tri12 homologue was 2.92-fold increased. Interestingly, F. oxysporum strain (10 L) showing similar growth patterns to the F. proliferatum isolates also indicated analogous Tri12 expression (3.06-fold–96 h after toxin exposition) (Table 2). Specificity of the transcription induction of Tri12b homologue genes in F. oxysporum (trichothecene transporter) was confirmed by the profiling of Tri12 gene expression in the presence of the multiple chemical compounds some of which can be potentially transported by the broad specific efflux pumps (Table 3). Only addition of deoxynivalenol significantly increased expression of Tri12 gene (13.45-fold). In the case of the rest used substances, addition of potentially harmful ferulic acid, coumaric acid, fungicide (Alert 375 SC), caffeine, H2O2 caused significant (P < 0.05) decrease of transcript level. Addition of sugars (glucose, sucrose), microelements (MgCl2, KCl) and potential host tissues (wheat leaves and roots) did not influence gene expression.
Transcriptional response of Tri201 homologues in F. oxysporum and F. proliferatum
To analyse general response of F. oxysporum and F. proliferatum strains to the presence of deoxynivalenol in the environment, expression of Tri201 gene was tested (Tri201 is a homologue of Tri101gene—responsible for detoxification by 3-O-acetylation of the trichothecene skeleton in the biosynthetic pathway in F. graminearum). In the presence of mycotoxin, Tri201 gene in F. oxysporum strains has shown significantly (P ≤ 0.05) increased transcriptional activity (13.07-fold–96 h after toxin exposition). No induction was observed in deoxynivalenol-treatedF. proliferatum isolates.
Sequence and phylogeny of Tri12 homologues
The protein sequences of Tri12 cluster members were clearly alignable with conserved transmembrane regions and (to a degree—Figs. 1 and 2) conserved splice junction positions in relation to the multiple sequence alignments.
Alignment of protein sequences arising from the translation of divergent Tri12 homologues shows conserved structural features. Grey rectangles correspond to consensus transmembrane region predictions, as predicted by TOPCONS and CCTOP. Blue arrows mark the conserved positions of splicing sites (based on gene models from Ensembl/Genbank corresponding nucleotide sequences). The poorly alignable N- and C-terminal regions were truncated. Sequences shown: PISL3812_09436—Talaromyces islandicus Tri12 homologue; FGSG_03541—F. graminearum Tri12; FsTRI12—F. sporotrichioides Tri12 (NCBI: AAK33071); TaTRI12—T. arundinaceum Tri12 (NCBI: CAY87358); TbTRI12—T. brevicompactum Tri12 (NCBI: CCA31154); SNOG_00428—Stagonospora nodorum Tri12 homologue; FFUJ_06844—F. fujikuroi Tri12 homologue; FproTRI12b—consensus protein sequence of Tri12 homologues from the analysed F. proliferatum isolates; FoxyTRI12b—consensus protein sequence of the Tri12 homologues from the analysed F. oxysporum isolates; FOPG_13450—Fusarium oxysporum f. sp. conglutinans Tri12 homologue
Extended majority rule consensus tree (maximum likelihood reconstruction) for non-redundant set of Tri12 homologues from different species (clustered at 97% protein sequence identity with CD-HIT, with exception of Fp and FoTRI12b sequences which represent consensus sequences for examined isolates). Scale is in amino acid residue change per site. Intron-exon structure is visualised (WebScipio remapping of original protein and nucleotide sequences). The tree was created with IQTREE v 1.3.6 (multi-threaded) using ultrafast bootstrap with automated stopping criterion based on topology convergence. The analysis was run with LG + G + F model (auto-selected). Following abbreviations were used for sequences obtained from NCBI: TaTRI12 (Trichoderma arundinaceum, CAY87358), TbTRI12 (T. brevicompactum, CCA31154), FsTRI12 (Fusarium sporotrichioides, AAK33071), FaeTRI12 (F. aethiopicum, ACJ69853), FcTRI12 (F. culmorum, AAM48786), FgTRI12 (F. graminearum, BAA76934), FmTRI12 (F. meridionale, AAM48906), FdTRI12 (F. dactylidis, AJC98152), other sequences were obtained from Ensembl/Fungi (v28) and are listed under their respective locus locus designations
The subsequent phylogenetic reconstruction of evolutionary relationships between Tri12 homologues (Fig. 2) has confirmed the distant relationship between canonical Tri12 genes present in sambucinum complex fusaria and more distant homologues (referred to as Tri12b) found in oxysporum and fujikuroi complexes. Majority of bipartitions were strongly supported (> 70% support) in ultrafast bootstrap analysis.
However, the attempts to root the resulting trees with even more distant homologues from DHA14 subset of MFS1 transporters have led to inconsistent results. This is likely due to ‘twilight zone’ (Rost 1999) levels of sequence similarity (around 20% protein sequence identity; BLAST expect values < 1e-20) in comparison to the considered outgroups (the sequences from 2.A.1.3 level of TCDB classification of transporters, e.g. Mfs1 from T. harzianum, Vba5p from Saccharomyces cerevisiae). Thus, we have opted for midpoint rooting in our reconstruction of the Tri12 ancestry (Hess and De Moraes Russo 2007).
Nucleotide sequence comparisons between the F. oxysporum and F. proliferatum isolates have shown their monophyleticity as members of their respective species complexes (Fig. 3—maximum likelihood tree). The sequence alignments have also uncovered a 25-26 bp indel differentiating between F. oxysporum and F. fujikuroi/proliferatum promotor regions (Fig. 4). We posit that this difference is possibly tied to the observed divergence in expressional patterns (see the following section for details). Interestingly, in F. oxysporum strain (10 L) showing similar growth patterns to the F. proliferatum isolates, sequence alignments have also uncovered a similar indel (Fig. 4). All Tri12 homologue sequences are available in NCBI database (KX273324, KX273325, KX273326, KX273327, KX273328, KX273329, KX273330, KX273331, KX273332, KX273333, KX273334, KX273335, KX273336, KX273337, KX273338, KX273339, KX273340, KX273341).
Majority rule consensus tree for Tri12b nucleotide sequences of examined F. oxysporum and F. proliferatum isolates (maximum likelihood reconstruction; bipartititions with support less than 50% were collapsed). Scale is in nucleotide changes per site. Both exon and intron sequences were used in the reconstruction—total of 1849 aligned positions. F. sporotrichioides Tri12 was used as outgroup to root the tree (not shown). The tree was created with IQTREE v 1.3.6 (multi-threaded) using ultrafast bootstrap with automated stopping criterion based on topology convergence. The analysis was run in partitioned mode, with separate models predicted for each exon and intron (respectively, exon 1–K2P + G, exon 2–TPM3 + G, exon 3–K2P + I, intron 1–K3P, intron 2–K2P). Additional Tri12 homologues from model genomes shown: FOQG_14213–F. oxysporum f. sp. raphani, FOTG_08309–F. oxysporum f. sp. vasinfectum, FOPG_13450–F. oxysporum f. sp. conglutinans, FOXB_15698–F. oxysporum Fo5176 (isolated from Arabidopsis thaliana), FFUJ_06844–F. fujikuroi
Alignment of pregenic sequence (ca. 100 base pairs) of examined F. oxysporum (Fo) and F. proliferatum (Fp) isolates shows a conserved motif (associated with lower gene expression) present in F. proliferatum and a single, early diverging F. oxysporum isolate (10 L). The ordering and included sequences from model genomes are same as on the Fig. 3
Discussion
Since the initial discovery and experimental characterisation of Tri12 efflux pomp (Alexander et al. 1999), subsequent inquiries have established its role in self-protection and virulence of trichothecene-producing strains (Menke et al. 2012). However, while the transformative detoxification mechanism in the form of Tri101O-3-acetyltransferase was found to be crucial in trichothecene resistance in multiple producing and non-producing species, the deletion experiments pointed to less significant role of the active efflux (Kimura et al. 2007; Khatibi et al. 2011). The presence and possible involvement of Tri12 homologues in non-producing species were largely left uninvestigated.
Our phylogeny reconstruction results (see Fig. 2) support early divergence of canonical Tri12 homologues in the Fusarium genus (present mostly in the sambucinum complex of the genus, as well as Trichoderma arundinaceum and T. brevicompactum) and the putative trichothecene transporter Tri12b (present in the oxysporum and fujikuroi complexes, with a notable exception of F. verticillioides—see Figs. 2 and 3). The phylogenetic placement of TaTri12 and TbTri12 points to a split predating the divergence of both genii and presence of additional homologues in Dothideomycetes (Diplodia seriata, Neofusicoccum parvum, Stagonospora nodorum) and Eurotiomycetes (Talaromyces islandicus) possibly substantiates either an even more distant relationship or possible spread via horizontal gene transfer.
During the preparation of the final stages of this work, a comprehensive analysis of the evolutionary origins of structural diversity in trichothecenes was put forth by Proctor and colleagues (Proctor et al. 2018). Based on multiple venues of evidence (trichothecene biosynthesis/metabolism-related phylogenies, chemical analyses and functional genomics evidence), the authors have strongly corroborated the diverse and discontinuous distribution of trichothecene biosynthetic capability among ascomycetes (in particular divergent Hypocreales species, as well as the incertae sedis ascomycete-Microcyclospora tardicrescens). The different scenarios implied by branching of gene histories in Nectriaceae, Cordycipitaceae and Hypocreaceae families underscore the possibility of both ancestral duplications and horizontal transfer between diverged donors/acceptors. Notably, in regards to TRI12, the conclusions have highlighted both optional role of the transporter in self-resistance of producers and possibility of compensatory role of different transporters with overlapping affinities for toxic compounds.
While the effects of trichothecenes on plant (Ohsato et al. 2007; Walter et al. 2010) and bacterial (Bisht et al. 2013) growth were previously characterised, the exact effect of the toxin on the growth of non-producing fusaria was, to our knowledge, previously not quantified. Through growth assays, we found that on the average the effects of trichothecene toxins on other fusaria are slight but statistically significant. On the average, Fusarium oxysporum isolates were found to be more resistant than F. proliferatum, with an exception of F. oxysporum 10 L. We found that the variation in resistance could likely be attributed to differences in the TRI12b homologue promoter region and overall transcriptional response (see also the following sections).
Our analysis of expression patterns (see Table 3) shows that F. oxysporum undergoes rapid shift in Tri12b expression upon trichothecene treatment. This reaction is not as strong in F. proliferatum and a divergent F. oxysporum isolate 10 L. Since no such response was observed on treatment with multiple different stressors (those agents that caused decreased growth also reduced expression of Tri12 probably due to the overall effect on fungal metabolism), we conclude that it should be considered as indirect evidence for Tri12b involvement, as an efflux pump contributing to the observed F. oxysporum resistance to trichothecene toxins. Additionally in the presence of mycotoxin, Tri201 gene (Tri201 is a homologue of Tri101gene—responsible for detoxification by 3-O-acetylation of the trichothecene skeleton in the biosynthetic pathway in F. graminearum) in F. oxysporum strains has also shown increased transcriptional activity whereas no induction was observed in deoxynivalenol treated F. proliferatum isolates.
The analysis of the promoter region of F. proliferatum and F. oxysporum isolates shows that this difference could possibly arise from a 25–26 bp deletion observed across all other oxysporum isolates. The cross-referencing with JASPAR-FUNGI database of known regulatory motifs shows that this stretch encodes possible pH-related TF binding site capable of binding PACC/RIM101 transcription factor (Supplementary Table 1). Deletion in this case would serve to decouple transporter expression from indirect environmental cues in form of pH (beneficial for a resistant non-producer) which has been previously documented to influence the expression of genes found in the canonical trichothecene cluster (Merhej et al. 2011) and virulence (Caracuel et al. 2003).
Notably, both predicted protein (aligned TM regions—Fig. 1) and gene structures (two introns of conserved position in regards to protein sequence—Figs. 1 and 2) are heavily conserved between both fusarial clades. Genes with rapid fluctuations are known to possess fewer exons, due to energetic costs associated with multiplicity of splicing events (Jeffares et al. 2008). As a toxin-associated efflux pump involved in early response Tri12b would thus be well tailored for rapid transcriptional response observed in the isolates of F. oxysporum.
References
Alexander NJ, McCormick SP, Hohn TM (1999) TRI12, a trichothecene efflux pump from Fusarium sporotrichioides: gene isolation and expression in yeast. Mol Gen Genet 261:977–984. https://doi.org/10.1007/s004380051046
Alexander NJ, McCormick SP, Hohn TM (2002) The identification of the Saccharomyces cerevisiae gene AYT1 (ORF-YLL063c) encoding an acetyltransferase. Yeast 19:1425–1430. https://doi.org/10.1002/yea.924
Bin-Umer MA, McLaughlin JE, Basu D, McCormick S, Tumer NE (2011) Trichothecene mycotoxins inhibit mitochondrial translation - implication for the mechanism of toxicity. Toxins 3:1484–1501. https://doi.org/10.3390/toxins3121484
Bisht SS, Praveen B, Amrita P, Santosh B, Kumar PK, Mishra R, Kanta PS (2013) Comparative study of various mycotoxins against few bacterial test organisms. J Pharm Res 10:116–119. https://doi.org/10.18579/jpcrkc/2011/10/3/89005
Caracuel Z, Roncero MI, Espeso EA, González-Verdejo CI, García-Maceira FI, Di Pietro A (2003) The pH signalling transcription factor PacC controls virulence in the plant pathogen Fusarium oxysporum. Mol Microbiol 48:765–779. https://doi.org/10.1046/j.1365-2958.2003.03465.x
Cardoza RE, Malmierca MG, Hermosa MR, Alexander NJ, McCormick SP, Proctor RH, Tijerino AM, Rumbero A, Monte E, Gutiérrez S (2011) Identification of loci and functional characterization of trichothecene biosynthesis genes in filamentous fungi of the genus Trichoderma. Appl Environ Microbiol 77:4867–4877. https://doi.org/10.1128/AEM.00595-11
Chang JM, Di Tommaso P, Notredame C (2014) TCS: a new multiple sequence alignment reliability measure to estimate alignment accuracy and improve phylogenetic tree reconstruction. Mol Biol Evol 3:1625–1637. https://doi.org/10.1093/molbev/msu117
Dawidziuk A, Koczyk G, Popiel D, Kaczmarek J, Buśko M (2014) Molecular diagnostics on the toxigenic potential of Fusarium spp. plant pathogens. J Appl Microbiol 116:1607–1620. https://doi.org/10.1111/jam.12488.24575830
Dawidziuk A, Koczyk G, Popiel D (2016) Adaptation and response to mycotoxin presence in pathogen- pathogen interactions within the Fusarium genus. World Mycotoxin J 9:565–575. https://doi.org/10.3920/WMJ2015.2010
Desjardins AE, Proctor RH (2007) Molecular biology of Fusarium mycotoxins. Int J Food Microbiol 119:47–50. https://doi.org/10.1016/j.ijfoodmicro.2007.07.024
Dobson L, Reményi I, Tusnády GE (2015) CCTOP: a consensus constrained TOPology prediction web server. Nucleic Acids Res 43:W408–W412. https://doi.org/10.1093/nar/gkv451
Ehrenhofer-Murray AE, Keller Seitz MU, Sengstag C (1998) The Sge1 protein of Saccharomyces cerevisiae is a membrane-associated multidrug transporter. Yeast 14:49–65. https://doi.org/10.1002/(SICI)1097-0061(19980115)14:1<49::AID-YEA199>3.0.CO;2-T
Frickey T, Lupas A (2004) CLANS: a Java application for visualizing protein families based on pairwise similarity. Bioinformatics 20:3702–3704. https://doi.org/10.1093/bioinformatics/bth444
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152. https://doi.org/10.1093/bioinformatics/bts565
Gromadzka K, Chelkowski J, Popiel D, Kachlicki P, Kostecki M, Goliński P (2009) Solid substrate bioassay to evaluate the effect of Trichoderma and Clonostachys on the production of zearalenone by Fusarium species. World Mycotoxin J 2:45–52. https://doi.org/10.3920/WMJ2008.x046
Hatje K, Keller O, Hammesfahr B, Pillmann H, Waack S, Kollmar M (2011)Cross-species protein sequence and gene structure prediction with fine-tuned Webscipio 2.0 and Scipio. BMC Res Notes 4:265. https://doi.org/10.1186/1756-0500-4-265
Hess PN, De Moraes Russo CA (2007) An empirical test of the midpoint rooting method. Biol J Linn Soc 92:669–674. https://doi.org/10.1111/j.1095-8312.2007.00864.x
Huerta-Cepas J, Dopazo J, Gabaldón T (2010) ETE: a python environment for tree exploration. BMC Bioinformatics 11:24. https://doi.org/10.1186/1471-2105-11-24
Jeffares DC, Penkett CJ, Bähler J (2008) Rapidly regulated genes are intron poor. Trends Genet 24:375–378. https://doi.org/10.1016/j.tig.2008.05.006
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Khatibi PA, Newmister SA, Rayment I, McCormick SP, Alexander NJ, Schmale DG (2011) Bioprospecting for trichothecene 3-O-acetyltransferases in the fungal genus Fusarium yields functional enzymes with different abilities to modify the mycotoxin deoxynivalenol. Appl Environ Microbiol 77:1162–1170. https://doi.org/10.1128/AEM.01738-10
Kimura M, Tokai T, Matsumoto G, Fujimura M, Hamamoto H, Yoneyama K, Shibata T, Yamaguchi I (2003) Trichothecene nonproducer Gibberella species have both functional and nonfunctional 3-O-acetyltransferase genes. Genetics 163:677–684
Kimura M, Tokai T, Takahashi-Ando N, Ohsato S, Fujimura M (2007) Molecular and genetic studies of Fusarium trichothecene biosynthesis: pathways, genes, and evolution. Biosci Biotechnol Biochem 71:2105–2123. https://doi.org/10.1271/bbb.70183
Koczyk G, Dawidziuk A, Popiel D (2015) The distant siblings - a phylogenomic roadmap illuminates the origins of extant diversity in fungal aromatic polyketide biosynthesis. Genome Biol Evol 7:3132–3154. https://doi.org/10.1093/gbe/evv204
Lee T, Lee SH, Shin JY, Yun JC, Lee YW, Ryu JG (2011) Occurrence of Fusarium mycotoxins in rice and its milling by-products in Korea. J Food Prot 74:1169–1174. https://doi.org/10.4315/0362-028X.JFP-10-564
Losada L, Ajayi O, Frisvad JC, Yu J, Nierman WC (2009) Effect of competition on the production and activity of secondary metabolites in Aspergillus species. Med Mycol 47:88–96. https://doi.org/10.1080/13693780802409542
Lutz MP, Feichtinger G, Défago G, Duffy B (2003) Mycotoxigenic Fusarium and deoxynivalenol production repress chitinase gene expression in the biocontrol agent Trichoderma atroviride P1. Appl Environ Microbiol 69:3077–3084. https://doi.org/10.1128/AEM.69.6.3077-3084.2003
Mathelier A, Fornes O, Arenillas DJ, Chen CY, Denay G, Lee J, Shi W, Shyr C, Tan G, Worsley-Hunt R, Zhang AW, Parcy F, Lenhard B, Sandelin A, Wasserman WW (2015) JASPAR 2016: a major expansion and update of the open-access database of transcription factor binding profiles. Nucleic Acids Res 44:D110–D115. https://doi.org/10.1093/nar/gkv1176
McLaughlin JE, Bin-Umer MA, Tortora A, Mendez N, McCormick S, Tumer NE (2009) A genome-wide screen in Saccharomyces cerevisiae reveals a critical role for the mitochondria in the toxicity of a trichothecene mycotoxin. Proc Natl Acad Sci 106:21883–21888. https://doi.org/10.1073/pnas.0909777106
Menke J, Yanhong D, Corby KH (2012)Fusarium graminearum Tri12p influences virulence to wheat and trichothecene accumulation. Mol Plant-Microbe Interact 25:1408–1418. https://doi.org/10.1094/MPMI-04-12-0081-R
Merhej J, Richard-Forget F, Barreau C (2011) The pH regulatory factor Pac1 regulates Tri gene expression and trichothecene production in Fusarium graminearum. Fungal Genet Biol 48:275–284. https://doi.org/10.1016/j.fgb.2010.11.008
Minh BQ, Nguyen MA, von Haeseler A (2013) Ultrafast approximation for phylogenetic bootstrap. Mol Biol Evol 30:1188–1195. https://doi.org/10.1093/molbev/mst024
Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ (2015) IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. https://doi.org/10.1093/molbev/msu300
O'Donnell K, Rooney AP, Proctor RH, Brown DW, McCormick SP, Ward TJ, Frandsen RJ, Lysøe E, Rehner SA, Aoki T, Robert VA, Crous PW, Groenewald JZ, Kang S, Geiser DM (2013) Phylogenetic analyses of RPB1 and RPB2 support a middle cretaceous origin for a clade comprising all agriculturally and medically important fusaria. Fungal Genet Biol 52:20–31. https://doi.org/10.1016/j.fgb.2012.12.004
Ohsato S, Ochiai-Fukuda T, Nishiuchi T, Takahashi-Ando N, Koizumi S, Hamamoto H, Kudo T, Yamaguchi I, Kimura M (2007) Transgenic rice plants expressing trichothecene 3-O-acetyltransferase show resistance to the Fusarium phytotoxin deoxynivalenol. Plant Cell Rep 26:531–538. https://doi.org/10.1007/s00299-006-0251-1
Pelletier B, Beaudoin J, Philpott CC, Labbé S (2003) Fep1 represses expression of the fission yeast Schizosaccharomyces pombesiderophore-iron transport system. Nucleic Acids Res 31:4332–4344. https://doi.org/10.1093/nar/gkg647
Perlin MH, Andrews J, Toh SS (2014) Essential letters in the fungal alphabet: ABC and MFS transporters and their roles in survival and pathogenicity. Adv Genet 85:201–253. https://doi.org/10.1016/B978-0-12-800271-1.00004-4
Picot A, Hourcade-Marcolla D, Barreau C, Pinson-Gadais L, Caron D, Richard-Forget F, Lannou C (2012) Interactions between Fusarium verticillioides and Fusarium graminearum in maize ears and consequences for fungal development and mycotoxin accumulation. Plant Pathol 61:140–151. https://doi.org/10.1111/j.1365-3059.2011.02503.x
Popiel D, Koczyk G, Dawidziuk A, Gromadzka K, Blaszczyk L, Chelkowski J (2014) Zearalenone lactonohydrolase activity in Hypocreales and its evolutionary relationships within the epoxide hydrolase subset of a/b-hydrolases. BMC Microbiol 14:82. https://doi.org/10.1186/1471-2180-14-82
Proctor RH, McCormick SP, Alexander NJ, Desjardins AE (2009) Evidence that a secondary metabolic biosynthetic gene cluster has grown by gene relocation during evolution of the filamentous fungus Fusarium. Mol Microbiol 74:1128–1142. https://doi.org/10.1111/j.1365-2958.2009.06927.x
Proctor RH, McCormick SP, Kim H-S, Cardoza RE, Stanley AM, Lindo L, Kelly A, Brown DW, Lee T, Vaughan MM, Alexander NJ, Busman M, Gutiérrez S (2018) Evolution of structural diversity of trichothecenes, a family of toxins produced by plant pathogenic and entomopathogenic fungi. PLoS Pathog 14(4):e1006946. https://doi.org/10.1371/journal.ppat.1006946
Ramakrishna N, Lacey J, Smith J (1996) The effects of fungal competition on colonization of barley grain by Fusarium sporotrichioides on T-2 toxin formation. Food Addit Contam 13:939–948. https://doi.org/10.1080/02652039609374481
Rost B (1999) Twilight zone of protein sequence alignments. Protein Eng 12:85–94. https://doi.org/10.1093/protein/12.2.85
Saier MH Jr, Reddy VS, Tsu BV, Ahmed MS, Li C, Moreno-Hagelsieb G (2016) The transporter classification database (TCDB): recent advances. Nucleic Acids Res 44:D372–D379. https://doi.org/10.1093/nar/gkv1103
Semeiks J, Borek D, Otwinowski Z, Grishin NV (2014) Comparative genome sequencing reveals chemotype-specific gene clusters in the toxigenic black mold Stachybotrys. BMC Genomics 15:1. https://doi.org/10.1186/1471-2164-15-590
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. https://doi.org/10.1093/molbev/mst197
Tokai T, Fujimura M, Inoue H, Aoki T, Ohta K, Shibata T, Yamaguchi I, Kimura M (2005) Concordant evolution of trichothecene 3-O-acetyltransferase and an rDNA species phylogeny of trichothecene-producing and non-producing fusaria and other ascomycetous fungi. Microbiology 151:509–519. https://doi.org/10.1099/mic.0.27435-0
Trapp SC, Hohn TM, McCormick S, Jarvis BB (1998) Characterization of the gene cluster for biosynthesis of macrocyclic trichothecenes in Myrothecium roridum. Mol Gen Genet 257:421–432. https://doi.org/10.1007/s004380050666
Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A (2015) The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 43:W401–W407. https://doi.org/10.1093/nar/gkv485
Walter S, Nicholson P, Doohan MF (2010) Action and reaction of host and pathogen during Fusarium head blight disease. New Phytol 185:54–66. https://doi.org/10.1111/j.1469-8137.2009.03041.x
Wuchiyama J, Kimura M, Yamaguchi I (2000) A Trichothecene efflux pump encoded by Tri102 in the biosynthetic gene cluster of Fusarium graminearum. J Antibiot 53:196–200. https://doi.org/10.7164/antibiotics.53.196
Acknowledgements
Research funded by National Science Centre, Poland, under SONATA research grant: “Molecular mechanisms of multidrug resistance to synthetic fungicides in fungi of the Fusarium genus” UMO-2011/03/D/NZ9/02061.
Funding
This study was funded by National Science Centre, Poland, under SONATA research grant: “Molecular mechanisms of multidrug resistance to synthetic fungicides in fungi of the Fusarium genus” UMO-2011/03/D/NZ9/02061.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
Delfina Popiel declares that she has no conflict of interest.
Adam Dawidziuk declares that he has no conflict of interest.
Grzegorz Koczyk declares that he has no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Additional information
Communicated by: Agnieszka Szalewska-Palasz
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOC 236 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Popiel, D., Dawidziuk, A. & Koczyk, G. Efflux pumps as an additional source of resistance to trichothecenes in Fusarium proliferatum and Fusarium oxysporum isolates. J Appl Genetics 60, 405–416 (2019). https://doi.org/10.1007/s13353-019-00501-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-019-00501-2