Abstract
The aim of this study was to predict the genomic breeding value (DGV) of production, selected conformation and reproductive traits, and somatic cell score of dairy cattle in Poland using high-frequency marker haplotypes. The dataset consisted of phenotypic, genotypic, and pedigree data of 1216 Polish Holstein-Friesian bulls. The genotypic data consisted of 54,000 single-nucleotide polymorphisms (SNPs). The data were divided into two subsets: a test dataset (n = 1064) and a validation dataset (n = 152). Genotypic data were selected using three criteria: the percentage of missing genotypes, minor allele frequency, and linkage disequilibrium. The purpose of the data selection was to identify blocks of SNPs that were then used for the construction of haplotypes. Only haplotypes with a frequency higher than 25% were selected. DGV was predicted using four variants of a linear model with random haplotype effects and deregressed breeding values as the response variables. The accuracy of genomic prediction was checked by comparing DGVs with estimated breeding values (EBVs) using two methods: Pearson’s correlations and the regression of EBV on DGV. The use of high-frequency haplotypes showed a tendency to underestimate DGVs. None of the models tested was clearly superior with regard to the traits studied. DGVs of production and conformation traits as well as somatic cell score (medium or high heritability traits) were more accurate than those estimated for fertility traits (low heritability traits).
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The genomic selection introduced by Meuwissen et al. (2001) has received much attention in animal breeding, because it provides predictions of the breeding values at a young age of animals with higher accuracy than breeding values based on parent average (Hess et al. 2017; Van Raden 2008). The potential benefits of using genomic selection were outlined by Schaeffer (2006). He showed that the genetic progress of a selected population of Canadian Holstein cattle would be doubled compared to the selection based on offspring phenotypes. He also stated that the use of genomic selection would reduce financial costs by 92% compared to traditional selection. Other benefits of genomic selection are decreased generation interval and the possibility of identifying recessive lethals (Wiggans et al. 2017). Furthermore, due to the lower production cost, a much larger number of bulls can be selected, which leads to a better management of genetic resources and limitation of inbreeding trends (Boichard et al. 2016).
The genomic breeding values (GBVs) in dairy cattle are usually predicted using single SNPs. However, a haplotype approach to the prediction of genomic breeding values using high density data is an alternative to single-marker methods (Calus et al. 2008; Cuyabano et al. 2014; Jónás et al. 2016; Hess et al. 2017). Building haplotypes based on linkage disequilibrium reduces the number of variables without the loss of information (Cuyabano et al. 2014).
An important benefit of haplotypes over SNP markers is their superior ability to identify mutations (Cuyabano et al. 2014). In addition, the use of haplotypes in genomic selection is advantageous because this approach treats the haplotype as a functional unit that contains the combined effects of tightly linked cis-acting causal variants (Da 2015; Garnier et al. 2013). On the other hand, the main disadvantage of models with marker haplotypes is that the number of effects to be estimated is significantly larger than that for SNP models (Calus et al. 2008). There are over a million haplotype alleles for a block of 20 biallelic SNPs, many of which occur at a low frequency (Hess et al. 2017). However, the number of haplotype effects to be estimated can be reduced by including more SNPs per haplotype or by using only haplotypes with a high frequency in the population. Discarding rare haplotype alleles also reduces computation time with little expected decrease in prediction accuracy (Gianola 2013). Other effective methods that reduce the number of explanatory variables in the linear model are, for example, the use of linkage disequilibrium (LD) to determine where a haplotype starts and ends in the genome (Gabriel et al. 2002), or the definition of haplotypes by setting windows with a fixed number of SNPs to form a haplotype (Villumsen et al. 2008).
The main purpose of this study was to predict the genomic breeding values of the production, selected conformation and reproductive traits, as well as somatic cell scores of Polish Holstein-Friesian cattle using high-frequency marker haplotypes. The study was divided into three stages: (1) selection of SNPs and blocks of linked markers, (2) construction of haplotypes and estimation of their frequency in the population under scrutiny, and (3) prediction of genomic breeding values using high-frequency haplotypes and comparison of the linear models applied.
Material and methods
Data
The dataset included the phenotype, genotype, and pedigree data of 1216 bulls of the Polish Holstein-Friesian breed. All the animals were born between 1987 and 2003. The most numerous group were born between 1997 and 2003 (n = 1061). The distribution of the number of bulls analyzed with respect to the year of their birth is shown in Fig. 1.
To compare prediction models most often so-called a K-fold cross-validation (K-fold CV) is used (Gianola and Schön 2016). However, in the present study, the CV layout was based on a generational partition of the dataset into two subsets (Perez-Cabal et al. 2012). One was the test dataset consisting of 1064 of the oldest individuals (87.5% of all the bulls) to train the model; the other contained the remaining 152 youngest individuals—this was the validation dataset. It has been shown (Daetwyler et al. 2008; Erbe et al. 2010) that larger predictive correlations in genomic selection are proportional to training sample size, thus, to increase predictive ability of the model, the test dataset was seven times larger than the validation dataset.
The study involved the following traits: milk yield (MY), fat yield (FY), protein yield (PY), stature (STA), overall feet and leg score (OFL), udder support (USU), non-return rate of cows (NRK), rest before pregnancy (PRP), time between pregnancies (OMC), and somatic cell score (SCS). MY, FY, and PY were measured in kilograms, while STA, OFL, and USU were included in the linear assessment expressed as points awarded (from 50 to 100 points for ST A and OFL, from 1 to 9 points for USU). NRK was 1 if a cow did not show symptoms of oestrus within 56 days of the first insemination, and 0 otherwise. PRP and OMC were expressed in days. SCS was calculated by converting the somatic cell count (SCC) using the following formula:
Estimated breeding values (EBVs) were obtained using a test day model with random regressions (Strabel et al. 2005). Heritabilities and variance components of the traits analyzed were estimated within the framework of the national genetic evaluation system of Polish Holstein-Friesian cattle (Interbull 2012). Their values are listed in Table 1.
Genotypic data were obtained using Illumina BovineSNP50 BeadChip (Matukumalli et al. 2009). The use of microarrays enabled the analysis of 54,001 SNPs for each individual. Out of 54,001 SNPs, 46,267 SNPs were selected for the DGV estimation based on their minor allele frequency (≥ 1%) and call rate (≥ 90%). This set of SNPs was subjected to further selection (the SNP selection criteria are described in the next subsection).
Statistical analysis
The statistical analysis was divided into three main steps: (1) selection of SNPs and blocks of linked markers, (2) construction of haplotypes and estimation of their frequency in the population analyzed, and (3) prediction of genomic breeding values using haplotypes and comparison of the linear models applied.
The first step in the SNP selection was to remove from the dataset those markers for which the missing data (genotypes) represented more than 10% of the total (GENO < 0.1). Then, the minor allele frequency (MAF) was used. Two limiting values were considered: 1 and 5%. As a result of this selection, two subsets of data were created: one with MAF > 0.01 and the other with MAF > 0.05. Based on the data subsets created in the previous step, blocks of linked SNPs were selected. LD of SNPs was measured using r2. Two limiting values were used: r2 ≥ 0.8 and r2 ≥ 0.9. Within each chromosome, r2 was calculated for each pair of SNPs. Linked markers were combined into blocks. As a result, four subsets of data were prepared: (1) MAF > 0.01 and r2 ≥ 0.8, (2) MAF > 0.01 and r2 ≥ 0.9, (3) MAF > 0.05 and r2 ≥ 0.8, and (4) MAF > 0.05 and r2 ≥ 0.9. SNP selection was carried out using the PLINK program (Purcell et al. 2007; Purcell 2010). Owing to the largest variety of block sizes, the first of the abovementioned subsets (MAF > 0.01 and r2 ≥ 0.8) was selected for further analysis. The blocks of SNPs were then used for the construction of haplotypes. This stage of the study, together with the estimation of haplotype frequency in the population, was done using the PHASE program (Stephens et al. 2001). In order to substantially reduce the number of explanatory variables in the linear models, only haplotypes with a frequency higher than 25% were selected for further analysis.
The genomic breeding values were predicted using four variants of the following linear model for each trait analyzed:
where y is the vector of deregressed breeding values, μ is the overall mean, Z is the design matrix for the random haplotype effects, h is the vector of the random haplotype effects, and ε is the vector of random errors, where ε ~ N (0, \( {\sigma}_e^2 \)) and \( {\sigma}_{\mathrm{e}}^2 \) is the error variance. The EBVs were deregressed using the method of Jairath et al. (1998).
Two variants of the above linear model (model 1 and model 2) used genetic variance equally divided into all haplotypes (regardless of their length), where \( h\sim N\left(0,\frac{\sigma_g^2}{\ \mathrm{number}\ \mathrm{of}\ \mathrm{haplotypes}}\right) \), \( \mathrm{and}\ {\sigma}_g^2 \) is the genetic variance of the trait. Furthermore, two types of the Z matrix were employed: model 1 used the Z matrix containing the probabilities of a given haplotype being passed on by a particular bull, while model 2 used the Z matrix consisting of only ones, if a given bull had a specific haplotype, and of zeros if it did not. Two other variants of the linear model (model 3 with the Z matrix defined as in model 1, and model 4 with the Z matrix defined as in model 2) used genetic variance unequally divided into all haplotypes (the larger the haplotype, the greater the part of the genetic variance assigned to it), where \( h\sim N\left(0,{\sigma}_g^2\bullet \frac{\mathrm{haplotype}\ \mathrm{size}}{\mathrm{number}\ \mathrm{of}\ \mathrm{alleles}}\right) \).
The genomic breeding value of the ith individual was estimated as follows: DGVi = ∑jZijhj.
The accuracy of the prediction of DGV was checked using two methods: Pearson’s correlations between EBV and DGV and the regression of EBV on DGV (Meuwissen et al. 2001).
All statistical analyses (unless otherwise stated) were performed using the R project (R Core Team 2011).
Results
Constructing haplotypes and estimating their frequency in the population
Haplotypes were constructed on the basis of the blocks selected—those selected from SNPs fulfilling the criteria of MAF > 0.01 and r2 ≥ 0.8 were considered the best set for haplotype construction. The reason for such a decision was the large variety of block sizes, especially the fact that the largest blocks were created in this subset. Table 2 shows the numbers of haplotypes constructed for individual chromosomes. The largest number of haplotypes was obtained for chromosome 26 (3709 haplotypes), whereas the smallest number of haplotypes was constructed for the sex chromosome (99 haplotypes) and for chromosome 28 (137 haplotypes). Also, chromosomes 23 and 24 were distinguished by the number of haplotypes, where their number exceeded 2000 (2069 and 2843, respectively). The number of haplotypes was dependent on block size. The larger the block, the more possible combinations of alleles, and thus the more possible haplotypes. The criterion for the selection of haplotypes was their frequency in the population. Only haplotypes which appeared in at least 25% of the individuals in the population were taken into account. Ultimately, the number of haplotypes was reduced from over 21,000 to 5682.
Predicting genomic breeding values
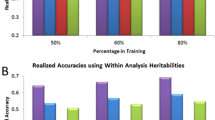
Tables 3 and 4 show the linear correlation coefficients between DGV and EBV for the test and validation datasets, respectively. In the test dataset, the strongest correlations were estimated for STA, FY, MY, PY, USU, and SCS, whereas lower correlations were estimated for reproductive traits (NRK, PRP, OMC). The linear correlation coefficients estimated in the validation dataset were lower; however, as in the test dataset, stronger correlations were estimated for medium and high heritability traits, while for the low heritability traits the estimated correlations were lower.
Tables 5 and 6 show regression coefficients of EBV on DGV for the test and validation datasets. All the tested models underestimated genomic breeding values. In the test dataset, the regression coefficients closest to the desired value (though a few times higher) were obtained for models 3 and 4. Models 1 and 2 displayed an evident tendency to underestimate breeding values, which was manifested by regression coefficients significantly exceeding unity. This tendency was particularly pronounced for NRK, PRP, and OMC (low heritability traits), and also for OFL and USU.
The analysis of EBV on DGV regression coefficients estimated for the validation dataset showed that models 1, 2, 3, and 4 maintained a tendency to underestimate breeding values, but the values of all the regression coefficients except MY decreased slightly. None of the linear models tested was clearly superior with regard to the studied traits.
Summing up, none of the linear models exhibited a significantly higher prediction accuracy in any of the comparisons between EBV and DGV, no matter whether correlations or regression were used: different types of design matrix for the random haplotype effects and the genetic variance equally or unequally divided into haplotypes had very little impact on the accuracy of prediction.
Discussion
Interest in haplotypes as explanatory variables in linear models for predicting genomic breeding values was generated almost parallel to the possibility of using information derived from SNPs (Calus et al. 2008; Tzeng and Bondell 2010; Jiang et al. 2012; Cuyabano et al. 2014; Jónás et al. 2016; Hess et al. 2017). Haplotypes can be constructed using various methods based on, for example, the expectation-maximization algorithm (Excoffier and Montgomery 1995) or the Bayesian theory (Stephens et al. 2001). In this study, based on a previous investigation (Macierzyńska and Wierzbicki 2008), the latter method was used.
The number of haplotypes constructed for the purposes of this study depended inter alia on the size of blocks. The more SNPs in the block, the more potential allele configurations in the haplotype. Owing to the large number of haplotypes constructed (explanatory variables), among which there were many with a low probability of occurrence in the population, the criterion of their frequency (> 25%) in the population was used. A similar approach was used by Huang et al. (2007), who used the limiting value of this criterion computed as 1/n and 2/n, where n is the population size. Also, Kolbehdari et al. (2007) and Boleckova et al. (2012) showed that predictions of breeding values using haplotypes with higher frequencies were more accurate than using haplotypes with lower frequencies. In the study by Hayes et al. (2006), haplotypes with a frequency of less than 1% were eliminated. That frequency was much lower than the haplotype frequency used in the present study. This was because of the large number of haplotypes constructed and the limited number of individuals analyzed, which caused problems with the linear modeling. According to Calus et al. (2008), reducing the number of haplotypes may improve both the feasibility and the power of the model.
The correlations between EBV and DGV obtained for the test dataset were high and comparable with those reported by Szyda et al. (2009), who used linear models with SNPs as random explanatory variables. The correlations estimated for the validation dataset were much lower. This tendency was also indicated by Habier et al. (2007) and Moser et al. (2009). They reported that a higher accuracy of prediction was achieved for the validation dataset using models with random haplotype effects. Boleckova et al. (2012) drew a similar conclusion after having tested linear models with haplotypes as explanatory variables. A like tendency was described by Mucha and Wierzbicki (2012), who used simulated data in a haplotype-based breeding value prediction study. The correlations between EBV and DGV for production traits in the validation dataset were lower than the correlations presented by Solberg et al. (2008). The authors reported correlations from 0.69 to 0.86 using SNPs, and from 0.80 to 0.82 using haplotypes. Kolbehdari et al. (2007) and Calus et al. (2009) also reported higher correlations between EBV and DGV: they ranged from 0.72 to 0.85 and from 0.79 to 0.81, respectively. However, it should be noted that all these studies were carried out using simulated data.
According to Hess et al. (2017), who studied the accuracy of genomic selection in dairy cattle using fixed-length haplotypes, fitting covariates for fixed-length haplotype alleles rather than SNPs can increase the accuracy of genomic prediction by up to 5.5%. They also found that using shorter haploblocks led to a higher accuracy of prediction than using longer ones.
The data used in the present study were also used to predict the breeding values based on SNP effects (Szyda et al. 2011). The reference population in that study was a group of 984 bulls, 252 of which belonged to the validation set. The correlations between EBV and DGV were 0.38 for milk yield, 0.37 for protein yield, and 0.32 for fat yield. These values were lower than the correlations estimated using models with random haplotype effects described in the present study. Szyda et al. (2011) estimated different correlation coefficients (0.43 for milk yield, 0.44 for protein yield, 0.31 for fat yield) after having estimated the genomic enhanced breeding value (GEBV) by combining DGV and parental information (parent average). The correlations between GEBV and EBV for milk yield and protein yield were comparable to the correlations between DGV and EBV reported in the present study.
The regression coefficients of EBV on DGV for the validation dataset indicated underestimation of GBVs of the studied traits. This was particularly evident with regard to low heritability traits (NRK, PRP, OMC). The least biased predictors were obtained for STA, MY, PY, FY, and SCS. This confirms that more accurate predictors of breeding values are obtained for traits with higher heritability (Calus et al. 2008; Moser et al. 2010). In Ireland, genomic selection was also implemented with a relatively small reference population of 596 bulls, and a validation group of 207 bulls (Berry and Kearney 2009). EBV on DGV regression coefficients were at the level of 0.76 for milk yield, 0.78 for fat yield, 0.80 for protein yield, and 0.77 for somatic cell score, which suggests that the predictors of breeding values, despite the differences between the reference population sizes, were more accurate than the predictors obtained in this study. However, Berry and Kearney (2009) pointed out that the accuracy of the results could have been greater if information from the bull daughters from the validation dataset had been included in the test dataset.
An interesting proposal regarding genomic selection using haplotypes was given by Da (2015). That author developed a quantitative genetics-based multi-allelic haplotype model for integrating functional and structural genomic information using haplotypes separately or jointly with SNPs. This approach may be a significant contribution to improving the accuracy of genomic prediction.
In the present study, a generational partition of the dataset into two subsets to perform cross-validation was applied. However, it is known that the CV layouts may have an important effect on the accuracy of genomic predictions (Perez-Cabal et al. 2012; Gianola and Schön 2016). The results of the study reported by Pszczola et al. (2012) indicate that genetic relationships within and between training and testing datasets influence the reliability of direct genomic breeding values. A higher relationship of the evaluated animals to the reference population together with smaller average relationship within the reference population results in higher reliabilities of genomic predictions. Keeping this in mind, further studies using different cross-validation scenarios would be needed to draw more general conclusions from the results of the study presented.
In conclusion, the use of high-frequency haplotypes showed a tendency to underestimate DGVs. None of the models tested was clearly superior with regard to the traits studied. The type of design matrix for the haplotype effects (either containing the probabilities of a given haplotype being passed on by a particular bull or consisting of only ones, if a given bull had a specific haplotype, and of zeros if it did not) as well as the equal or unequal division of the genetic variance between haplotypes did not significantly affect the accuracy of prediction. DGVs of production and conformation traits as well as somatic cell score (medium or high heritability traits) were more accurate than those estimated for fertility traits (low heritability traits). The relatively low accuracy of genomic selection, especially for low heritability traits, could well have been higher if high-frequency haplotypes had been combined with SNPs alleles with known effects (Calus et al. 2008) or if haplotype effects had been combined with parental information (Szyda et al. 2011).
References
Berry D, Kearney F (2009) Genomic selection in Ireland. Proceedings of the Interbull International Workshop - Genomic Information in Genetic Evaluations, Uppsala, Sweden, Bulletin No. 39:29-34.
Boichard D, Ducrocq V, Croiseau P, Fritz S (2016) Genomic selection in domestic animals: principles, applications and perspectives. C R Biologies 339:274–277
Boleckova J, Christensen OF, Sorensen P, Sahana G (2012) Strategies for haplotype association mapping in a complex pedigreed population. Czech J Anim Sci 57:1–9
Calus MPL, Meuwissen THE, de Roos APW, Veerkamp RF (2008) Accuracy of genomic selection using different methods to define haplotypes. Genetics 178:553–561
Calus MPL, Meuwissen THE, Windig JJ, Knol EF, Schrooten C, Vereijken ALJ, Veerkamp RF (2009) Effects of the number of markers per haplotype and clustering of haplotypes on the accuracy of QTL mapping and prediction of genomic breeding values. Genet Select Evol 41:11
Cuyabano BCD, Su G, Lund MS (2014) Genomic prediction of genetic merit using LD-based haplotypes in the Nordic Holstein population. BMC Genomics 15(1):1171
Da Y (2015) Multi-allelic haplotype model based on genetic partition for genomic prediction and variance component estimation using SNP markers. BMC Genet 16:144
Daetwyler HD, Villanueva B, Woolliams JA (2008) Accuracy of predicting the genetic risk of disease using a genome-wide approach. PLoS One 3:e3395
Erbe M, Pimentel ECG, Sharifi AR, Simianer H (2010) Assessment of cross-validation strategies for genomic prediction in cattle. Book of Abstracts of the 9th World Congress of Genetics Applied to Livestock Production, Leipzig, Germany, p. 129
Excoffier L, Montgomery S (1995) Maximum-likelihood estimation of molecular haploype frequencies in a diploid population. Mol Biol Evol 12(5):921–927
Gabriel SB, Schaffner SF, Nguyen H, Moore JM, Roy J, Blumenstiel B, Higgins J, DeFelice M, Lochner A, Faggart M, Liu-Cordero SN, Rotimi C, Adeyemo A, Cooper R, Ward R, Lander ES, Daly MJ, Altshuler D (2002) The structure of haplotype blocks in the human genome. Science 296:2225–2229
Garnier S, Truong V, Brocheton J, Zeller T, Rovital M, Wild PS et al (2013) Genome-wide haplotype analysis of cis expression quantitative trait loci in monocytes. PLoS Genet 9(1):e1003240
Gianola D (2013) Priors in whole-genome regression: the Bayesian alphabet returns. Genetics 194:573–596
Gianola D, Schön CC (2016) Cross-validation without doing cross-validation in genome enabled prediction. G3 6(10):3107–3128
Habier D, Fernando RL, Dekkers JCM (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177:2389–2397
Hayes B, Hagesaether N, Adnoy T, Pellerud G, Berg PR, Lien S (2006) Effects on production traits of haplotypes among casein genes in Norwegian goats and evidence for a site of preferential recombination. Genetics 174:455–464
Hess M, Druet T, Hess A, Garrick D (2017) Fixed-length haplotypes can improve genomic prediction accuracy in an admixed dairy cattle population. Genet Select Evol 49:54
Huang BE, Amos CI, Lin DY (2007) Detecting haplotype effects in genomewide association studies. Genet Epidemiol 31:803–812
Interbull (2012). http://www.interbull.org. Accessed 12 March 2012
Jairath L, Dekkers JCM, Schaeffer LR, Liu Z, Burnside EB, Kolstad B (1998) Genetic evaluation for herd life in Canada. J Dairy Sci 81:550–562
Jiang J, Tomohiro N, Masanori S, Naoyuki S, Aoi N, Mikano S, Yoichi I, Kasamaki Y, Masakatsu O, Masayoshi S, Koichi M, Hiroshi K, Yukio O, Ma Y (2012) Haplotype of smoothelin gene associated with essential hypertension. Hereditas 149:178–185
Jónás D, Ducrocq V, Fouilloux MN, Croiseau P (2016) Alternative haplotype construction methods for genomic evaluation. J Dairy Sci 99(6):4537–4546
Kolbehdari D, Schaeffer LR, Robinson JAB (2007) Estimation of genome wide haplotype effects in half-sib designs. J Anim Breed Genet 124(6):356–361
Macierzyńska A, Wierzbicki H (2008) Statistical aspects of selecting informative SNPs and estimating haplotype frequencies. J Agrobiol 25(1):31–34
Matukumalli LK, Lawley CT, Schnabel RD, Taylor JF, Allan MF, Heaton MP et al (2009) Development and characterization of a high density SNP genotyping assay for cattle. PLoS One 4:e5350
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157:1819–1829
Moser G, Tier B, Crump RE, Khatkar MS, Raadsma HW (2009) A comparison of five methods to predict genomic breeding values of dairy bulls from genome-wide SNP markers. Genet Select Evol 31(41):56
Moser G, Khatkar M, Hayes B, Raadsma H (2010) Accuracy of direct genomic values in Holstein cows using subsets of SNP markers. Proceedings of the 9th World Congress on Genetics Applied to Livestock Production, 1–6 August 2010, Leipzig, Germany
Mucha A, Wierzbicki H (2012) Linear models for breeding values prediction in haplotype-assisted selection. BMC Proc 6(Suppl 2):S11
Perez-Cabal MA, Vazques AI, Gianola D, Rosa GJM, Weigel KA (2012) Accuracy of genome enabled prediction in a dairy cattle population using different cross-validation layouts. Front Genet 3:27
Pszczola M, Strabel T, Mulder HA, Calus MP (2012) Reliability of direct genomic values for animals with different relationships within and to the reference population. J Dairy Sci 95(1):389–400
Purcell S (2010) PLINK (1.07). Documentation. http://zzz.bwh.harvard.edu/plink/dist/plink-doc-1.07.pdf. Accessed 1 March 2012
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, deBakker PIW, Daly MJ, Sham PC (2007) Plink: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet 81:559–575
R Core Team (2011) R: A language and environment for statistical computing. Vienna, Austria : the R Foundation for Statistical Computing. ISBN: 3-900051-07-0. Available online at http://www.R-project.org/. Accessed 12 March 2012
Schaeffer LR (2006) Strategy for applying genome-wide selection in dairy cattle. J Anim Breed Genet 123(4):218–223
Solberg TR, Sonesson AK, Woolliams JA, Meuwissen THE (2008) Genomic selection using different marker types and densities. J Anim Sci 86(10):2447–2454
Stephens M, Smith N, Donnelly P (2001) A new statistical method for haplotype reconstruction from population data. Am J Hum Genet 68:978–989
Strabel T, Szyda J, Ptak E, Jamrozik J (2005) Comparison of random regression test-day models for Polish Black and White cattle. J Dairy Sci 88:3688–3699
Szyda J, Kamiński S, Żarnecki A, Żukowski K (2009) Incorporation of correlation between SNPs into the genomic evaluation model. Proceedings of the Interbull Meeting in Barcelona, Spain No 40:193–197
Szyda J, Żarnecki A, Suchocki T, Kamiński S (2011) Fitting and validating the genomic evaluation model to Polish Holstein-Fresian cattle. J Appl Genet 52:363–366
Tzeng J, Bondell HD (2010) A comprehensive approach to haplotype-specific analysis by penalized likelihood. Eur J Human Genet 18:95–103
Van Raden PM (2008) Efficient methods to compute genomic predictions. J Dairy Sci 91:4414–4423
Villumsen TM, Janss L, Lund MS (2008) The importance of haplotype length and heritability using genomic selection in dairy cattle. J Anim Breed Genet 126:3–13
Wiggans GR, Cole JB, Hubbard SM, Sonstegard TS (2017) Genomic selection in dairy cattle: the USDA experience. Annu Rev Anim Biosci 5:309–327
Acknowledgements
We are very grateful to Anna Ruść and Elżbieta Wójcik for technical assistance. The entire dataset (phenotype, genotype, and pedigree data as well as EBVs and deregressed EBVs of the individuals studied) was provided by the MASinBULL Consortium.
Author information
Authors and Affiliations
Contributions
Anna Mucha designed the study, performed the statistical analysis, interpreted results, and wrote the manuscript. Heliodor Wierzbicki designed the study, interpreted results, and wrote the manuscript. Stanisław Kamiński supervised laboratory work. Kamil Oleński performed laboratory work. Dorota Hering performed laboratory work.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by: Maciej Szydlowski
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Mucha, A., Wierzbicki, H., Kamiński, S. et al. High-frequency marker haplotypes in the genomic selection of dairy cattle. J Appl Genetics 60, 179–186 (2019). https://doi.org/10.1007/s13353-019-00489-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-019-00489-9