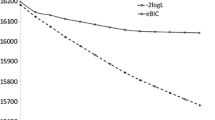
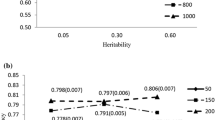
Abstract
The objective was to evaluate the effects of directional selection based on estimated genomic breeding values (GEBVs) for a quantitative trait. Selection affects GEBV prediction accuracy as well as genetic architecture via changes in allelic frequencies and linkage disequilibrium (LD), and the resulting changes are different from those in the absence of selection. How marker density affects long-term GEBV accuracy and selection response needs to be understood as well. Simulations were used to characterize the impact of selection based on GEBVs over generations. Single-nucleotide polymorphism (SNP) marker effects were estimated with the Bayesian Lasso method in the base generation, and these estimates were used to calculate the GEBVs in subsequent generations. GEBV accuracy decreased over generations of selection, and it was lower than under random selection, where a decay took place as well. In the long term, selection response tended to reach a plateau, but, at higher marker density, both the magnitude and duration of the response were larger. Selection changed quantitative trait loci (QTL) allele frequencies and generated new but unfavorable LD for prediction. Family effects had a considerable contribution to GEBV accuracy in early generations of selection.











Similar content being viewed by others
References
Abasht B, Sandford E, Arango J, Settar P, Fulton JE, O’Sullivan NP, Hassen A, Habier D, Fernando RL, Dekkers JC, Lamont SJ (2009) Extent and consistency of linkage disequilibrium and identification of DNA markers for production and egg quality traits in commercial layer chicken populations. BMC Genomics 10(Suppl 2):S2
Bulmer MG (1971) The effect of selection on genetic variability. Am Nat 105(943):201–211
Crow JF (2008) Maintaining evolvability. J Genet 87:349–353
Daetwyler HD, Pong-Wong R, Villanueva B, Woolliams JA (2010) The impact of genetic architecture on genome-wide evaluation methods. Genetics 185(3):1021–1031
de los Campos G, Naya H, Gianola D, Crossa J, Legarra A, Manfredi E, Weigel KA, Cotes JM (2009) Predicting quantitative traits with regression models for dense molecular markers and pedigree. Genetics 182(1):375–385
Dekkers JCM, van Arendonk JAM (1998) Optimizing selection for quantitative traits with information on an identified locus in outbred populations. Genet Res 71(3):257–275
Fernando RL (2009) Genomic selection: Bayesian methods. Animal Breeding & Genetics Short Courses, Iowa State University, Ames, IA
Gianola D, de los Campos G, Hill WG, Manfredi E, Fernando RL (2009) Additive genetic variability and the Bayesian alphabet. Genetics 183:347–363
Gibson JP (1994) Short-term gain at the expense of long-term response with selection on identified loci. In: Proceedings of the 5th World Congress on Genetics Applied to Livestock Production, Guelph, Canada, 7–12 August 1994, pp 201–204
Goddard M (2009) Genomic selection: prediction of accuracy and maximisation of long term response. Genetica 136(2):245–257
Habier D, Fernando RL, Dekkers JC (2007) The impact of genetic relationship information on genome-assisted breeding values. Genetics 177(4):2389–2397
Hill WG (2010) Understanding and using quantitative genetic variation. Philos Trans R Soc Lond B Biol Sci 365(1537):73–85
Jannink J-L (2010) Dynamics of long-term genomic selection. Genet Sel Evol 42:35
Meuwissen T, Goddard M (2010) Accurate prediction of genetic values for complex traits by whole-genome resequencing. Genetics 185(2):623–631
Meuwissen THE, Hayes BJ, Goddard ME (2001) Prediction of total genetic value using genome-wide dense marker maps. Genetics 157(4):1819–1829
Muir WM (2007) Comparison of genomic and traditional BLUP-estimated breeding value accuracy and selection response under alternative trait and genomic parameters. J Anim Breed Genet 124(6):342–355
Park T, Casella G (2008) The Bayesian Lasso. J Am Stat Assoc 103:681–686
Slatkin M (2008) Linkage disequilibrium—understanding the evolutionary past and mapping the medical future. Nat Rev Genet 9(6):477–485
Solberg TR, Sonesson AK, Woolliams JA, Meuwissen THE (2008) Genomic selection using different marker types and densities. J Anim Sci 86:2447–2454
Solberg TR, Sonesson AK, Woolliams JA, Odegard J, Meuwissen THE (2009) Persistence of accuracy of genome-wide breeding values over generations when including a polygenic effect. Genet Sel Evol 41:53
Sonesson AK, Meuwissen THE (2009) Testing strategies for genomic selection in aquaculture breeding programs. Genet Sel Evol 41:37
Spelman RJ, Garrick DL (1997) Utilisation of marker assisted selection in a commercial dairy cow population. Livest Prod Sci 47:139–147
Villanueva B, Dekkers JCM, Woolliams JA, Settar P (2004) Maximizing genetic gain over multiple generations with quantitative trait locus selection and control of inbreeding. J Anim Sci 82:1305–1314
Vukasinovic N, Moll J, Kunzi N (1998) Genetic gain from QTL selection versus traditional selection methods. In: Proceedings of the 49th Annual Meeting of the European Association for Animal Production, Warsaw, Poland, 24–27 August 1998
Wright S (1937) The distribution of gene frequencies in populations. Proc Natl Acad Sci USA 23:307–320
Yi N, Xu S (2008) Bayesian LASSO for quantitative trait loci mapping. Genetics 179(2):1045–1055
Acknowledgments
This work was supported by the Wisconsin Agriculture Experiment Station, and by grants NRICGP/USDA 2003-35205-12833, NSF DEB-0089742, and NSF DMS-044371. We thank the editor of the journal and the reviewers for their insightful comments.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Long, N., Gianola, D., Rosa, G.J.M. et al. Long-term impacts of genome-enabled selection. J Appl Genetics 52, 467–480 (2011). https://doi.org/10.1007/s13353-011-0053-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-011-0053-1