Abstract
A repetitive sequence of 491 bp, named pMD232-500, was isolated from S. cereale cv. Kustro using wheat SSR marker Xgwm232. GenBank BLAST search revealed that the sequence of pMD232-500 was highly similar to a part of retrotransposon Nusif-1. Fluorescence in situ hybridization (FISH) analysis using pMD232-500 as probe indicated that only 14 Thinopyrum intermedium chromosomes and all the chromosomes of S. cereale cv. Kustro bear FISH signals, however, no FISH signals were observed on Dasypyrum villosum chromosomes. In addition, the FISH signals were distributed on whole arms except their terminal regions. Further genomic in situ hybridization (GISH) analysis using genomic DNA from Pseudoroegneria spicata indicated that the 14 Th. intermedium chromosomes bearing FISH signals should belong to J genome. Thereafter, the repetitive elements pMD232-500 showed the unambiguous features of genomic constitution of Th. intermedium. In addition, the results in the present study have indicated the similarity of genomes from Th. intermedium and S. cereale.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
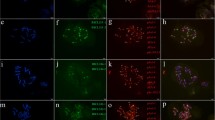
The tribe Triticeae provides a vast gene pool for most agronomically important traits including some that do not exist in wheat. The wild wheatgrass, Thinopyrum intermedium (Host) Barkworth and Dewey (= Agropyron intermedium (Host) P. B.) is a hexaploid species (2n = 6x = 42) with genomes E1E2St (Wang et al. 1994) or JJSSt (Chen et al. 1998). The species has been hybridized extensively with wheat and proved to be a valuable source of genes for disease resistance (Larkin et al. 1995; Chen et al. 2003; Yang et al. 2006; Luo et al. 2009) and tolerance to salt stress (Dvořák 1985). The correct identification of Th. intermedium chromosomes involved in these gene transfers is important for cytogenetic investigations. Genomic in situ hybridization (GISH) has been used to examine the genomic constitution of Th. intermedium (Chen et al. 1998) and the results indicated that the S genomic DNA probe from the diploid species Pseudoroegneria spicata can effectively separate the Th. intermedium chromosomes into J, JS and S (S = St) genomes (Chen et al. 1998; Chen 2005). However, S specific genomic DNA probe cannot divide J and JS genomes into two obvious groups, which contain the basic 14 chromosomes in Triticeae species (Chen et al. 1998). That is, the exact genomic constitution of Th. intermedium is very complex and still unsettled, and has showed ambiguous features (Kishii et al. 2005). A new Sabrina-like long terminal repeat (LTR) pDbH12 has been isolated from Dasypyrum breviaristatum and this repetitive element could serve as a cytogenetic marker for tracing JS genome in Th. intermedium (Liu et al. 2009). As so far, no repetitive elements have been obtained for tracing J genome of Th. intermedium. In the present study, a new repetitive DNA sequence belonging to a LTR-retrotransposon Nusif-1 family was isolated from S. cereale cv. Kustro, and its chromosomal distribution was identified by FISH. A wheat SSR marker Xgwm232 (Röder et al. 1998) was screened for amplification in T. aestivum cv. Mianyang11, T. aestivum cv. Chinese Spring (CS) and S. cereale cv. Kustro. This SSR marker gave rise to a specific DNA band of about 500 bp from S. cereale cv. Kustro and this band was not amplified from the two wheat cultivars (Fig. 1a). Subsequently, the rye-specific band was cloned and sequenced. The full length of the 491-bp sequence was obtained and designated as pMD232-500 (GenBank accession No. EF535858). BLAST search in the NCBI GenBank revealed that the 278-482 bp nucleotide sequence of pMD232-500 shows 77% identity to a LTR-retrotransposon Nusif-1 of T. aestivum (GenBank accession No. DQ537335). This shows that wheat SSR markers can be used to isolate the retrotransposon sequences in Triticeae plants enriching with repetitive elements. In order to determine the chromosomal distribution of the isolated LTR-retrotransposon like sequence, pMD232-500 was labeled with digoxigenin-11-dUTP by nick translocation, and the probe was used to hybridize the mitotic metaphases cells of S. cereale cv. Kustro, CS, Th. intermedium and D. villosum (PI 598390, supported by National Plant Germplasm System, USDA-ARS, Aberdeen, Idaho, USA) by FISH analysis with no block. The process was according to the protocols from Tang et al. (2008). All the chromosomes of S. cereale cv. Kustro and only 14 Th. intermedium chromosomes bear FISH signals (Fig. 1b-c), however, no FISH signals were observed on CS and D. villosum chromosomes (data not shown). In addition, the FISH signals were distributed on whole arms except their terminal regions (Fig. 1b-c). For determining the genome affiliations of the chromosomes bearing FISH signals in Th. intermedium, total genomic DNA from Ps. spicata was also labeled with digoxigenin-11-dUTP by nick translation, and the probe was used to hybridize the same mitotic metaphases cells of Th. intermedium, which have already been used for FISH analysis. The sheared genomic DNA of CS was used as blocking DNA. The process was according to the protocols described by Yang et al. (2006). In the Th. intermedium, the GISH analysis indicated that 14 chromosomes were strongly and uniformly hybridized along the entire chromosome length, eight chromosomes showed strong signals around the centromeres and weaker signals at the telomeres (Fig. 1d). According to the genomic formula of Th. intermedium created by Chen et al. (1998), the 14 chromosomes should belong to the St genome, the eight chromosomes belong to the JS genome, and the remaining 20 chromosomes belong to the J genome. Apparently, the pMD232-500 probe only hybridized to 14 of the 20 chromosomes. This result indicated that the 14 chromosomes bearing FISH signals of pMD232-500 probe should be an integrated set of genome chromosomes and they should be different from the other six of the 20 chromosomes. In the present study, six of the 20 chromosomes were putatively included into JS genome (Fig. 1d) and the 14 chromosomes bearing FISH signals were designated as J genome. Liu et al. (2009) have isolated a repetitive element pDbH12, which gives rise to hybridization signals on 14 Th. intermedium chromosomes and can be used to identify the JS genome. In addition, pMD232-500 probe was used to hybridize another root-tip preparation of Th. intermedium. After rinsing the root-tip preparation, FISH analysis using pDbH12 as a probe was carried out on the same slide. Results apparently indicated that pMD232-500 probe and pDbH12 probe hybridized to different groups of chromosome of Th. intermedium (Fig. 2). The pDbH12 sequence must be different from the pMD232-500 sequence because it was isolated from D. villosum and could hybridize to D. villosum chromosomes, however, the pMD232-500 was isolated from rye and could not hybridize to D. villosum chromosomes. Thereafter, pDbH12, pMD232-500 and genomic DNA from Ps. spicata can be used to identify JS, J and St genomes of Th. intermedium, respectively. Using these two repetitive elements and the genomic DNA from Ps. spicata, the exact genomic constitution of Th. intermedium has been settled and has shown unambiguous features.
PCR, FISH patterns of pMD232-500, and GISH pattern of the genomic DNA from Pseudoroegneria spicata: a PCR analysis with primer pair Xgwm232 in wheat and rye: 1 = S. cereale cv. Kustro; 2 = T. aestivum cv. Chinese Spring; 3 = T. aestivum cv. Mianyang11; M = DNA marker; arrow indicates the target band, b FISH pattern of metaphase chromosomes in S. cereale cv. Kustro, using pMD232-500 as probe, c and d Sequential FISH and GISH on the same spread of metaphase chromosomes of Th. intermedium, using pMD232-500 and genomic DNA from Ps. spicata as probes, respectively. The J, JS and St genomes have been marked
In the present study, we distinguished the J genome in Th. intermedium using FISH of pMD232-500 sequence. The hybridization pattern of the J genome was similar to R genome chromosomes of S. cereale. This result indicated similarity of genomes from Th. intermedium and S. cereale. Kishii et al. (2005) have tentatively re-designated the genomic formula of Th. intermedium as StJS(V-J-R)S, superscripted s stands for St genome signals in E/J and V-J-R genomes, and V-J-R stands for derivative genome involved in the evolution of the three genomes. This formula also implies that Th. intermedium has a close relationship with S. cereale. Liu et al. (2009) have modified the genome formula of Th. intermedium as StJ(V-J-H) S, superscripted s stands for St genome signals in V-J-H genomes, and V-J-H stands for derivative genome involved in the evolution of the three genomes. Thereafter, as that indicated by Liu et al. (2009), it is possible that an ancestral genome was (V-J-R-H) that evolved to (V-J-R) and (V-J-H), which in turn diverged further into (V-J), (V-R), (J-R), (V-H), and (J-H), prior to evolving to the diploids V, J, R and H.
In conclusion, the new LTR-retrotransposon-like sequence pMD232-500 can be used to identify J genome of Th. intermedium, and the further investigation of its distribution on the chromosomes of other Triticeae species is worth doing.
References
Chen Q (2005) Detection of alien chromatin introgression from Thinopyrum into wheat using S genomic DNA as a probe–A landmark approach for Thinopyrum genome research. Cytogenet Genome Res 109:350–359
Chen Q, Conner RL, Laroche A, Thomas JB (1998) Genome analysis of Thinopyrum intermedium and Th. ponticum using genomic in situ hybridization. Genome 41:580–586
Chen Q, Conner RL, Li HJ, Sun SC, Ahmad F, Laroche A, Graf RJ (2003) Molecular cytogenetic discrimination and reaction to wheat streak mosaic virus and the wheat curl mite in Zhong series of wheat–Thinopyrum intermedium partial amphiploids. Genome 46:135–145
Dewey DR, 1984. The genomic system of classification as a guide to intergeneric hybridization with the perennial Triticeae. In: Gustafson JP (ed.), Gene Manipulation in Plant Improvement. Plenum, New York 16:209-279.
Dvořák J (1985) Transfer of salt tolerance from Elytrigia pontica (Podp.) Holub. to wheat by the addition of an incomplete Elytrigia genome. Crop Sci 21:306–309
Kishii M, Wang RRC, Tsujimoto H, 2005. GISH analysis revealed new aspect of genomic constitution of Thinopyrum intermedium. In: Czech J. Genet. Plant Breed, eds. Proc. 5th Int Triticeae Symp, Prague, Czech Republic: 92-95.
Larkin PJ, Banks PM, Lagudah ES, Apple R, Chen X, Xin ZY, Ohm HW, McIntosh RA (1995) Disomic Thinopyrum intermedium addition lines in wheat with barley yellow dwarf virus resistance and with rust resistances. Genome 38:385–394
Liu C, Yang ZJ, Jia JQ, Li GR, Zhou JP, RTen ZL (2009) Genomic distribution of a long terminal repeat(LTR) Sabrina-like retrotransposon in Triticeae species. Cereal Res Commun 37:363–372
Luo PG, Luo HY, Chang ZJ, Zhang HY, Zhang M, Ren ZL (2009) Characterization and chromosomal location of Pm40 in common wheat: a new gene for resistance to powdery mildew derived from Elytrigia intermedium. Theor Appl Genet 118:1059–1064
Röder MS, Korzun V, Wendehake K, Plaschke J, Tixier MH, Leroy P, Ganal MW (1998) A microsatellite map of wheat. Genetics 149:2007–2023
Tang ZX, Fu SL, Ren ZL, Zhou JP, Yan BJ, Zhang HQ (2008) Variation of tandem repeat, regulatory element, and promoter regions revealed by wheat-rye amphiploids. Genome 51:399–408
Wang RRC, von Bothmer R, Dvorák J, Fedak G, Linde-Laursen I, Muramatsu M (1994) Genomic symbols in the Triticeae. In: Wang RRC, Jensen KB, Jaussi C (eds) Proc. 2nd Int Triticeae Symp, Logan, Utah. Utah State University Publication Design and Production, Logan, pp 29–34
Yang ZJ, Li GR, Chang ZJ, Zhou JP, Ren ZL (2006) Characterization of a partial amphiploid between Triticum aestivum cv. Chinese Spring and Thinopyrum intermedium ssp. trichophorum. Euphytica 149:11–17
Acknowledgements
We are grateful for financial support from the national Natural Science Foundation of China (grants no. 30730065 and 30871518), China Postdoctoral Science Foundation (grant no.20090450110) and National 973 Program (grant no. 2009CB118300).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Tang, Z.X., Yang, Z.J., Fu, S.L. et al. A new long terminal repeat (LTR) sequence allows to identify J genome from JS and St genomes of Thinopyrum intermedium . J Appl Genetics 52, 31–33 (2011). https://doi.org/10.1007/s13353-010-0019-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13353-010-0019-8