Abstract
Pineapple plants (hybrid MD2) with bacterial heart rot were detected in a commercial plantation at Glasshouse Mountains, Queensland, in November 2015. The bacterial strain BRIP64263 isolated from infected tissue was shown to be a Gram negative soft-rotting bacterium capable of growth at 41 ºC, and based on its culture properties was provisionally identified as Dickeya. This strain was compared with other putative Dickeya strains affecting banana (BRIP64262) and potato (BRIP29490). Sequence analysis of the recombinase A genes of the pineapple strain placed it in phylotype I of D. zeae, whereas the banana strain was placed in phylotype II. This was confirmed by sequence comparisons for the phosphofructose kinase, RNA polymerase and aconitase genes which showed that the pineapple strain BRIP64263 is distinct from other strains that infect pineapples and other hosts in Australia and overseas. Furthermore, phylogenetic analysis of the replication initiation factor gene showed that strains affecting pineapples were distributed among both phylotypes of D. zeae, indicating multiple acquisitions or opportunistic infections of pineapple from this group of pathogens. The potato isolate, BRIP29490, was shown to be Rahnella aquatica, and is not likely to be pathogenic. It is not known whether the new isolate represents an incursion or whether it has long been associated with pineapples in Australia. Further study is required to determine the epidemiological characteristics of this strain, and what threat it poses to Australian pineapple production.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Pineapple (Ananas comosus) is an iconic crop for Queensland, Australia. Originating in South America, it is grown commercially throughout tropical and subtropical regions of South America, the Pacific, Asia, Africa and Australia. Australian pineapple production is centred around Beerwah/Glasshouse mountains (southeast Queensland), Bundaberg and Yeppoon (east-central), and Rollingstone (northeast). Although a significant plantation was established in the Northern Territory, production has now ceased. Pineapples constitute a relatively small but valuable commodity for Australian horticulture, with an estimated annual value of over $AU 50 million ($USD 38 million) (Australian Horticulture Statistics Handbook 2020). In addition to its value as an agricultural commodity, pineapples have been identified as an iconic crop with significant value in tourism and branding.
The Australian pineapple industry is relatively free from pests and diseases. The major issue identified is root and/or heart rot caused by the oomycetes Phytophthora cinnamomi and P. nicotianae (Pegg 1977, Anderson et al. 2012). Losses from this disease have increased with the cultivation of the low acidity hybrids MD2 and 73 − 50. These hybrids also have greater propensity for natural flowering, and MD2 in particular is very susceptible to bacterial heart rot and fruit collapse. The Australian industry was previously based on Smooth Cayenne which has resistance to bacterial heart rot and fruit collapse. A bacterial heart rot of pineapple does occur in Queensland if dirty water is used when applying urea fertilizer. The urease in the water breaks urea down to NH4OH which damages the soft tissues of the heart allowing entry for unspecified strains of the bacterium formerly known as Erwinia chyrsanthemi (Rohrbach and Johnson 2003).
Bacterial heart rot of pineapple is caused by poorly-characterised bacterial strains belonging to the genus Dickeya (Samson et al. 2005). The causal agent was originally placed within Erwinia chrysanthemi, before being transferred to Pectobacterium (Brenner et al. 1977, Hauben et al. 1998). However, it is now considered to belong to a group of strains among Dickeya zeae (Samson et al. 2005). In addition to the transmission pathways of other bacterial soft-rots caused by subspecies of Pectobacterium carotovorum (that is, infected juice, wind and rain-spatter), bacterial heart rot can be transmitted by insects, such as ants, that visit the flowers (Lim and Lowings 1979). Thus, far from being an opportunistic infection, the bacterial strains that cause heart rot are highly adapted to flower transmission (Lim 1974).
Pineapple heart rot was first identified in Malaysia in 1927 (Lim 1974). The disease is first observed on the inner leaves as water soaked lesions that coalesce to form blisters as the bacteria ferment plant products. Eventually these will release infectious bacterial ooze. Infected fruit will rapidly collapse as it approaches maturity. The fermentation of fruit products leads to a build-up of gas. As the gas escapes it can make an audible hissing sound, particularly in the quiet of night. This led to its other name, ghost rot, as it frightened many field workers as they would return to their homes at night. In a 1976 address to the Australasian Plant Pathology Society, Lim Wen Hee stated that he had “even heard of farmers who had summoned witch doctors to chant prayers to exorcise spirits which seemed to have developed a taste for pineapples.”
Pineapple heart rot can result in significant yield losses of up to 40%. It has been reported from Malaysia (Lim and Lowings 1979), the Philippines, Indonesia, Costa Rica (Chinchilla et al. 1979), Brazil (Melo et al. 1974) and Hawaii (Kaneshiro et al. 2008). In November 2015, a commercial pineapple grower at Glasshouse Mountains, Queensland, noticed strange symptoms on two plants of the variety MD2. There was concern that the new strain may be the same that causes ghost rot in Hawaii and Malaysia (Lim and Lowings 1979; Kaneshiro et al. 2008).
In June 2010, an Import Risk Analysis (IRA) was commenced for the importation of fresh, decrowned pineapple fruit from Malaysia into Australia. At the time, the risk posed by what was termed ‘Erwinia chrysanthemi’ was considered ‘very low’ because Australia already had E. chrysanthemi infections of banana, potato and sugarcane. However, at that stage it was not known whether the endemic Australian strains of what was previously known as E. chrysanthemi could have any impact on pineapples. However, expert advice provided to Biosecurity Australia explained that nomenclature using ‘E. chrysanthmi’ was obsolete and that the Malaysian strains were distinct from Australian strains (Australian Senate Rural and Regional Affairs and Transport References Committee 2014). Upon review, no pineapples were imported. However, in November 2015, when the new disorder was detected in commercial pineapples, there was a requirement to identify the strain responsible in order to better understand the implications its presence presented to Australia’s biosecurity framework .
Materials and methods
Bacterial isolation and characterisation
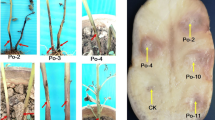
Pineapple plants of the MD2 cultivar exhibiting symptoms of heart rot (Fig. 1) were collected in November 2015 from a commercial plantation at Glass House Mountains in Southeast Queensland. These samples were sent to the Centre of Tropical Agriculture, Mareeba, Queensland, for processing. A bacterium, designated BRIP64263 (Mareeba collection = J4236), was isolated onto nutrient agar (NA) from symptomatic leaf and heart rot tissue. This bacterium was subject to standard bacteriological characterisation, including Gram stain, potassium hydroxide (3%) mucosity test, motility, potato soft rot test, growth at 37˚C and 41˚C, growth on CPG (Kelman 1954), King’s Medium B (King et al. 1954), and M3SC medium (Young et al. 2016). Additionally, a putative ‘Erwinia chrysanthemi’ BRIP64262 (Gatton collection = B1B2) isolated from banana in far north Queensland, and a putative ‘E. chrysanthemi’ BRIP29490 isolated from potatoes in Victoria, were subject to the same screening for comparative purposes. Strains used in this study are presented in Table 1.
Pathology testing
The pathogenicity of isolate BRIP64263 was determined by inoculating asymptomatic pineapple tops of three cultivars: MD2, Smooth Cayenne and 73 − 50. Cultures used for inoculation were grown for 48 h on NA medium, and a water suspension water adjusted to 108 CFU mL− 1 was prepared after scraping bacterial growth from the surface of the plates. Five leaves from each cultivar were detached and inoculated using the technique of Kaneshiro et al. (2008). Prior to inoculation, leaves were disinfected with 70% ethanol and were air dried at room temperature. Two sets of five leaves of each variety were inoculated with 0.5 mL (each leaf) of bacterial suspension. In one set a small piece of inoculum-saturated cotton was placed over the incision and held in place with transparent tape, however in other sets only bacterial suspensions were placed over incisions. Similar methods were followed in the control treatments, but sterilised distilled water was used for inoculations.
Inoculated leaves of each individual variety were placed in separate aluminium trays lined with filter paper moistened with sterile distilled water. The trays were covered with plastic bag for 24 h and incubated in a growth chamber at 33 ± 2 ºC and approximately 80% relative humidity. After 24 and 48 h, inoculation sites were observed for symptom development. Isolations were made from tan coloured water soaked lesions from selected inoculated leaves of each cultivar following standard bacterial isolation procedures.
Molecular characterisation
The major taxonomic work for Dickeya was conducted by Parkinson et al. (2009), and therefore the primers for the four loci used in that work were used for PCR. These loci were aconitase (acn), RNA polymerase (rpo), phosphofructokinase (pfk) and recombinase A (recA). Additionally, a search of the GenBank database of the National Center for Biotechnology Information (NCBI) revealed that multiple pineapple Dickeya strains had been examined using the DNA replication initiation factor locus (dnaA) (Schneider et al. 2011). Owing to technical issues described by Schneider et al. (2011), a total of four primer sets were ordered to ensure amplification and useful sequence data for this locus. The 16 S rRNA gene was also examined using the primers of Lane 1991. All primers used in this study are presented in Table 2.
PCR was conducted in 25 µL volumes and included 12.5 µL of 2x Amplitaq Gold 360 (Thermo Fisher Scientific), 0.5 µL of 10 µM of the forward and reverse primers for each respective locus, 10.5 µL of ddH2O and 1 µL of template. Bacterial templates were made by thoroughly mixing a small loop-full of bacterial colony growth into 1 mL of ddH2O. Initially, the following thermocycle was used: 95˚C for 5 min, 35 cycles of 95˚C for 30 s, 60˚C for 30 s and 72˚C for 30 s, followed by a final extension at 72˚C for 7 min. PCR products (5 µL) were visualised using GelRed stain incorporated in 1.5% agarose gels poured and run in 0.5x TAE buffer.
The putative Dickeya strains produced good products for the rpoD and pfk loci, but only weak products for the others. Therefore, for the remaining primer sets, the annealing temperature was amended to 51˚C, which facilitated amplification from all loci for BRIP64263. Products (20 µL) were purified and Sanger sequencing reactions were performed by Macrogen Inc, South Korea.
Results
Bacterial isolation and characterisation
Several basic tests were conducted as these can be useful in discriminating soft-rotting bacteria (Table 1). All strains were Gram negative, based on standard Gram staining and the potassium hydroxide mucosity test, and all were motile, as determined by observation of directional swimming of cells using phase contrast microscopy with 1,000x magnification. The pineapple and banana strains were both positive for the potato soft-rot test, although they produced distinct rotting properties, and both grew strongly at 41˚C. These properties are consistent with the group of bacteria formerly classified as E. chrysanthemi but now described as Dickeya. In contrast, the potato isolate did not cause soft rotting, and grew very weakly at 37˚C and failed to grow at 41˚C. These properties suggest that BRIP29490 is not a member of the soft-rotting group of bacteria, and as such has been mis-identified.
Pathology testing
Inoculation of BRIP64263 bacterial isolate on leaves of three commercial varieties of pineapple, MD2, Smooth Cayenne and 73 − 50, confirmed that the isolate is capable of inducing light tan to brown coloured water soaked symptoms after 48 h. These symptoms closely resembled those on affected MD2 plants from Glasshouse Mountains (Fig. 2). BRIP64263 was found to affect all three varieties and produced tan to brown coloured water soaked lesions on inoculated leaves, however they did not form a gas filled blister as has been reported for other pineapple heart rots. No symptoms were observed in control treatments. Reisolation of the same bacterium from inoculated leaves completed Koch’s postulates and confirmed it was the causal agent.
Molecular characterisation
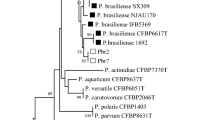
Sequences were generated for all 5 loci for each of the putative Dickeya strains, BRIP64263 and BRIP64262. Two loci, RNA polymerase and replication initiation factor, were obtained for BRIP29490. BLAST results indicated that BRIP64263 and BRIP64262 belonged to the Dickeya group of pathogens, while BRIP29490 gave high matches to Rahnella aquatilis, a non-pathogenic plant associate. Maximum Likelihood phylogenetic trees for the recA (Fig. 3) and dnaA (Fig. 4) loci were prepared by downloading relevant reference sequences and aligning them using MUSCLE in MEGAX (Kumar et al. 2018). Phylogenetic trees for the other loci could not be prepared owing to a lack of available reference sequences on GenBank. A General Time Reversible model was used with 1,000 bootstrap replications. The Subtree-Pruning-Regrafting ML heuristic method was employed after the initial tree was constructed using the Neighbour-Joining method. These loci were selected because of the availability of reference sequences of Dickeya strains known or suspected to cause ghost rot of pineapple. The pineapple pathogen, BRIP64263 could be assigned to Phylotype II in relation to classification studies conducted by Parkinson et al. (2009).
Maximum Likelihood phylogenetic tree (1,000 replications) inferred through 415 bp of DNA sequence from the recombinase A (recA) gene used by Parkinson et al. (2009) to review the genus. Statistics for topologies supported by 50% or more shown. Scale shows number of substitutions per 100 bp. Strains sequenced in this study are indicated in bold
Maximum Likelihood phylogenetic tree inferred through 700 bp of DNA sequence from the replication initiation factor locus, (dnaA) gene used by Schneider et al. (2011). Statistics for topologies supported by 50% or more shown. Scale shows number of substitutions per 100 bp. Dickeya strains associated with pineapple are highlighted, and strains sequenced in this study are indicated in bold
Discussion
This study identified a novel soft-rotting member of Dickeya zeae was responsible for heart rot symptoms of pineapple in Glass House Mountain, Queensland, Australia. Phylogenetic analysis of the recombinase A (recA) gene identified it as belonging to the Phylotype I group of soft-rotting bacteria identified by Parkinson et al. (2009) (Fig. 3). In addition to the recA analysis, BLAST analysis of the phosphofructose kinase, RNA polymerase and aconitase genes demonstrated it to be distinct from all other characterised strains (Table 3). These strains all belong to Dickeya, confirming the genus of the Australian isolate, but none gave a complete match for any of the loci sequenced. Furthermore, it is distinct from the banana corm rot organism, strain BRIP64262, that was previously identified as Erwinia chrysanthemi, but is placed in Phylotype II based on the recA results. A third strain (BRIP29490) isolated from potatoes and supplied as ‘E. chrysanthemi’ was actually shown to be a plant associate called Rahnella aquatilis. This strain tested negative for pectolytic activity in potato and is not believed to be pathogenic.
The extent to which the novel infection identified here is present through Australia’s pineapple industry is unknown. While the pathogen responsible is closely related to strains that cause pineapple heart rot in Malaysia, it is clearly different as the Malaysian pineapple strains belong to Phylotype II (Fig. 3). Interestingly, using the Australian pineapple (BRIP64263) and banana (BRIP64262) isolates as representatives of the different phylotypes, it is possible to further characterise a broader suite of pineapple-associated strains using the additional dnaA sequence data that is available for this group. This reveals that Dickeya zeae strains that infect pineapple are distributed among the two phylotypes (Fig. 4).
It is not known whether this strain represents an incursion or if it has been present for a long period of time. While BRIP64263 is clearly highly pathogenic on a range of pineapple cultivars (Fig. 2), there have been very few officially reported instances of the disease. It has been observed in Rollingstone, where incidence appeared to be associated with feeding wounds from a heavy infestation of red mite, and Yeppoon (Col Scott, pers. comm. 2017). The industry was previously based on Smooth Cayenne which was considered resistant to bacterial soft rots and rarely produces out-of-cycle flowers. However, the move into low acid hybrids has problems associated with their propensity for natural flowering and greater susceptibility to bacterial heart rot. It is possible that growers may not recognise or be hesitant to report the disease, or it could be that aspects of pineapple agronomy in Australia help mitigate transmission. For example, it is known that Malaysian strains of the pineapple heart rot pathogen can be transmitted by insects visiting the flower (Lim and Lowings 1979). As Australian growers force flowering with ethylene, most flowers appear and then disappear within a relatively short time period. In contrast, with natural flowering, an infected insect could seek out new flowers as they progressively appear, facilitating broader field-wide transmission.
It should be noted that bacterial heart rot is unlikely to be a natural disease of pineapple. The pineapple evolved in South America, but the disease originated in Malaysia, suggesting that this represents a new encounter disease. There must have been an original host in Malaysia from which the virulent strain jumped across to pineapple. Thus, while it is possible that the current D. zeae strain BRIP64263 arrived with pineapple germplasm during an unidentified period, it is also possible that it is an indigenous strain that has emerged in association with highly susceptible MD2 plants with their tendency to flower naturally.
The identification of a new and potentially damaging strain of D. zeae affecting pineapples has relevance to the broader interests of plant health in Australia and worldwide. Taxonomy is fluid, but decisions need to be based on the most current information available, and not rely on assumptions based on older nomenclatural arrangements (Young 2018). In the data presented here, the bacteria isolated from pineapple and banana belong to different phylotypes, and are likely to have significant differences in their epidemiology. The data highlight the importance of using the most current and accepted taxonomic nomenclature; old and defunct species identifications can result in poor import risk analyses. As shown in this study, strains whose microbial properties initially led them to be classified as E. chrysanthemi were clearly distinct from each other, and thus represented individual risks.
International trade is an important agency to improve world living standards. Within the International Plant Protection Convention (IPPC) and World Trade Organisation (WTO) framework, countries must make trade decisions based on scientifically sound advice. If a pathogen or pest is already present in the importing country, the commodity can no longer be subject to considerations involving that pathogen or pest unless it can be demonstrated to be an epidemiologically distinct strain. For example, wheat stem rust, caused by Puccinia graminis f. sp. tritici is already present in Australia, but the epidemiological distinct Ug99 strain poses a significant threat to Australia’s wheat crop. With the advent of rapid and cost-effective genome sequencing technologies, there is significant scope to screen strains from the importing and exporting countries where the nomenclature indicates the species is present in both regions. In time, this might involve identification of specific virulence factors that are responsible for different epidemiologies. With increasing pressures on global food supply, it is necessary to have an adaptive and consultative framework in order to mitigate against the spread of plant pathogens between regions.
References
Anderson JM, Pegg KG, Scott C, Drenth A (2012) Phosphonate applied as a pre-plant dip controls Phytophthora cinnamomi root and heart rot in susceptible pineapple hybrids. Austral Plant Pathol 41:59–68
Australian Horticulture Statistics Handbook (2019) HA18002-Australian-Horticulture-Statistics-Handbook-2019-20-Fruit.pdf
Australian Senate Rural and Regional Affairs and Transport References Committee (2014) Inquiry into the effect on Australian pineapple growers of importing fresh pineapples from Malaysia. ISBN 978-1-74229-986-0
Brenner DJ, Fanning GR, Steigerwalt AG (1977) Deoxyribonucleic acid relatedness among erwiniae and other enterobacteria. II. Corn stalk rot bacterium and Pectobacterium chrysanthemi. Int J Syst Bacteriol 27:211–221
Chinchilla CM, Gonzales LC, Morales F (1979) Bacterial heart rot (Erwinia chrysanthemi) of pineapple in Costa Rica. Agron Cost 3:183–185
Hauben L, Moore ERB, Vauterin L, Steenackers M, Mergaert J, Verdonck L, Swings J (1998) Phylogenetic position of phytopathogens within the Enterobacteriaceae. Syst Appl Microbiol 21:384–397
Kaneshiro WS, Burger M, Vine BG, de Silva AS, Alvarez AM (2008) Characterization of Erwinia chrysanthemi from a bacterial heart rot of pineapple outbreak in Hawaii. Plant Dis 92:1444–1450
Kelman A (1954) The relationship of pathogenicity in Pseudomonas solanacearum to colony appearance on a tetrazolium medium. Phytopath 44:693–695
King EO, Ward MK, Raney DE (1954) Two simple media for the demonstration of pyocyanin and fluoroscein. J Lab Clin Med 44:301–307
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: Molecular Evolutionary Genetics Analysis across computing platforms. Mol Biol and Evol 35:1547–1549
Lane DJ(1991) 16S/23S rRNA sequencing, p. 115–175.InE. Stackebrandtand M. Goodfellow (ed.), Nucleic acid techniques in bacterial systematics. J.Wiley & Sons, Ltd., Chichester, United Kingdom
Lim WH (1974) The etiology of fruit collapse and bacterial heart rot of pineapple. MARDI Res Bull 2:11–16
Lim WH, Lowings PH (1979) Pineapple fruit collapse in peninsular Malaysia: symptoms and varietal susceptibility. Plant Disease Reporter 63:170–174
Melo SAP, Araujo E, Suassauna J (1974) Preliminary note of the occurrence of Erwinia sp. causing a dry rot of pineapple fruits in Paraiba. Rev of Agric 49:124
Parkinson N, Stead D, Bew J, Heeney J, Tsror (Lahkim) L, Elphinstone J(2009) Dickeya species relatedness and clade structure determined by comparison of recA sequences. Int J of Syst and Evol Micro 59: 2388–2393
Pegg KG (1977) Soil application of elemental sulphur as a control of Phytophthora cinnamomi root and heart rot of pineapple. Aust J of Exp Agric and Animal Husbandry 17:859–865
Rohrbach KG, Johnson MW (2003) Pests, diseases and weeds. In: Bartholomew DP, Paull RE, Rohrbach KG (eds) The Pineapple: Botany, Production and Uses. CAB International, Wallingford, Oxfordshire, pp 203–251
Samson R, Legendre JB, Christen R, Fischer-Le Saux M, Achouak W, Gardan L(2005) Transfer of Pectobacterium chrysanthemi (Burkholder et al. 1953) Brenner et al. 1973 and Brenneria paradisiaca to the genus Dickeya gen. nov. as Dickeya chrysanthemi comb. nov. and Dickeya paradisiaca comb. nov. and delineation of four novel species, Dickeya dadantii sp. nov., Dickeya dianthicola sp. nov., Dickeya dieffenbachiae sp. nov. and Dickeya zeae sp. nov. Int J of Syst and Evol Micro 55: 1415–1427
Schneider KL, Marrero G, Alvarez AM, Presting GG (2011) Classification of plant associated bacteria using RIF, a computationally derived DNA marker. PLoS ONE 6(4):e18496. doi:https://doi.org/10.1371/journal.pone.0018496
Young AJ, Kawamata A, Ensbey M, Lambley E, Nock CJ (2016) Efficient diagnosis of ratoon stunting disease of sugarcane by quantitative PCR on pooled leaf sheath biopsies. Plant Disease 100: 2492–2498.
Young A (2018) The rough end of the pineapple: the sometimes prickly relationship between science and policy. International Congress of Plant Pathology (ICPP) 2018: Plant Health in A Global Economy. APSNET
Acknowledgements
The authors wish to acknowledge the late Colin Scott (Agronomist, Tropical Pines), who was one of the foremost advocates for the Australian pineapple industry, and who passed away in 2018. We also wish to acknowledge discussions we had with Toni Chapman (NSW Department of Primary Industries) and José Liberato (Biosecurity and Animal Welfare Division, Northern Territory Department of Primary Industry and Resources).
Funding
Open Access funding enabled and organized by CAUL and its Member Institutions
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Young, A.J., Pathania, N., Manners, A. et al. Heart rot of Australian pineapples caused by Dickeya zeae. Australasian Plant Pathol. 51, 525–533 (2022). https://doi.org/10.1007/s13313-022-00880-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13313-022-00880-x