Abstract
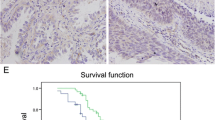
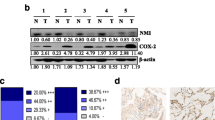
Lung cancer is still the leading cause of malignant deaths in the world. It is of great importance to find novel functional genes for the tumorigenesis of lung cancer. We demonstrated that Rac3 could promote cell proliferation and inhibit apoptosis in lung adenocarcinoma cell line A549 previously. The aim of this study was to investigate the function and mechanism of Rac3 in lung adenocarcinoma cell lines. Immunohistochemistry staining was performed in 107 lung adenocarcinoma tissues and matched non-tumor tissues. Multivariate analysis and Kaplan-Meier analysis were used to investigate the correlation between Rac3 expression and the clinical outcomes. 3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) assay, colony formation assay, and flow cytometry analysis were employed to determine the proliferative ability, cell cycle distribution, and apoptosis in H1299 and H1975 cell lines. Gene expression microarray and pathway analysis between the Rac3-siRNA group and the control group in A549 cells were performed to investigate the pathways and mechanism of Rac3 regulation. Rac3 was shown to be positively expressed in lung adenocarcinoma tissues, and the expression of Rac3 associates with longer survival in lung adenocarcinoma patients. Silencing of Rac3 significantly induced cell growth inhibition, colony formation decrease, cell cycle arrest, and apoptosis of lung adenocarcinoma cell lines, which accompanied by obvious downregulation of CCND1, MYC, and TFDP1 of cell cycle pathway involving in the tumorigenesis of lung adenocarcinoma based on the gene expression microarray. In conclusion, these findings suggest that Rac3 has the potential of being a therapeutic target for lung adenocarcinoma.






Similar content being viewed by others
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29.
GBD 2013 Mortality and Causes of Death Collaborators. Global, regional, and national age-sex specific all-cause and cause-specific mortality for 240 causes of death, 1990-2013: a systematic analysis for the global burden of disease study 2013. Lancet. 2015;385:117–71.
Reddy C, Chilla D, Boltax J. Lung cancer screening: a review of available data and current guidelines. Hosp Pract. 2011;39:107–12.
Bach PB, Mirkin JN, Oliver TK, Azzoli CG, Berry DA, Brawley OW, Byers T, Colditz GA, Gould MK, Jett JR, Sabichi AL, Smith-Bindman R, Wood DE, Qaseem A, Detterbeck FC. Benefits and harms of CT screening for lung cancer: a systematic review. JAMA. 2012;307:2418–29.
Botling J, Edlund K, Lohr M, Hellwig B, Holmberg L, Lambe M, Berglund A, Ekman S, Bergqvist M, Pontén F, König A, Fernandes O, Karlsson M, Helenius G, Karlsson C, Rahnenführer J, Hengstler JG, Micke P. Biomarker discovery in non–small cell lung cancer: integrating gene expression profiling, meta-analysis, and tissue microarray validation. Clin Cancer Res. 2013;19:194–204.
Pusztai L. Chips to bedside: incorporation of microarray data into clinical practice. Clin Cancer Res. 2006;12:7209–14.
Van Aelst L, D’Souza-Schorey C. Rho GTPases and signaling networks. Genes Dev. 1997;11:2295–322.
Rossman KL, Der CJ, Sondek J. GEF means go: turning on RHO GTPases with guanine nucleotide-exchange factors. Nat Rev Mol Cell Biol. 2005;6:167–80.
Sahai E, Marshall CJ. Rho-GTPases and cancer. Nat Rev Cancer. 2002;2:133–42.
Ridley AJ. Rho GTPase and cell migration. J Cell Sci. 2001;114:2713–22.
Fritz G, Just I, Kaina B. Rho GTPases are over-expressed in human tumors. Int J Cancer. 1999;81:682–7.
Abraham MT, Kuriakose MA, Sacks PG, Yee H, Chiriboga L, Bearer EL, Delacure MD. Motility-related proteins as markers for head and neck squamous cell cancer. Laryngoscope. 2001;111:1285–9.
Haataja L, Groffen J, Heisterkamp N. Characterization of RAC3, a novel member of the rho family. J Biol Chem. 1997;272:20384–8.
Mira JP, Benard V, Groffen J, Sanders LC, Knaus UG. Endogenous, hyperactive Rac3 controls proliferation of breast cancer cells by a p21-activated kinase-dependent pathway. Proc Natl Acad Sci U S A. 2000;97:185–9.
Gest C, Joimel U, Huang L, Pritchard LL, Petit A, Dulong C, Buquet C, Hu CQ, Mirshahi P, Laurent M, Fauvel-Lafève F, Cazin L, Vannier JP, Lu H, Soria J, Li H, Varin R, Soria C. Rac3 induces a molecular pathway triggering breast cancer cell aggressiveness: differences in MDA-MB-231 and MCF-7 breast cancer cell lines. BMC Cancer. 2013;13:63.
Walker MP, Zhang M, Le TP, Wu P, Lainé M, Greene GL. RAC3 is a pro-migratory co-activator of ERα. Oncogene. 2011;30:1984–94.
Engers R, Ziegler S, Mueller M, Walter A, Willers R, Gabbert HE. Prognostic relevance of increased Rac GTPase expression in prostate carcinomas. Endocr Relat Cancer. 2007;14:245–56.
Dong S, Zhao J, Wei J, Bowser RK, Khoo A, Liu Z, Luketich JD, Pennathur A, Ma H, Zhao Y. F-box protein complex FBXL19 regulates TGFβ1-induced E-cadherin down-regulation by mediating Rac3 ubiquitination and degradation. Mol Cancer. 2014;13:76.
Li J, Liu Y, Yin Y. Inhibitory effects of Arhgap6 on cervical carcinoma cells. Tumor Biol. 2015 1. [Epub ahead of print]
Liu TQ, Wang GB, Li ZJ, Tong XD, Liu HX. Silencing of Rac3 inhibits proliferation and induces apoptosis of human lung cancer cells. Asian Pac J Cancer Prev. 2015;16:3061–5.
Lois C, Hong EJ, Pease S, Brown EJ, Baltimore D. Germline transmission and tissue-specific expression of transgenes delivered by lentiviral vectors. Science. 2002;295:868–72.
Ginzinger DG. Gene quantification using real-time quantitative PCR: an emerging technology hits the mainstream. Exp Hematol. 2002;30:503–12.
Franken NA, Rodermond HM, Stap J, Haveman J, van Bree C. Clonogenic assay of cells in vitro. Nat Protoc. 2006;1:2315–9.
Milner AE, Levens JM, Gregory CD. Flow cytometric methods of analyzing apoptotic cells. Methods Mol Biol. 1998;80:347–54.
Allemani C, Weir HK, Carreira H, Harewood R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A, Marcos-Gragera R, Stiller C, Azevedo e Silva G, Chen WQ, Ogunbiyi OJ, Rachet B, Soeberg MJ, You H, Matsuda T, Bielska-Lasota M, Storm H, Tucker TC, Coleman MP. CONCORD working group. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (CONCORD-2). Lancet. 2015;385:977–1010.
Sánchez de Cos Escuín J. Molecular staging and prognosis in lung cancer. Arch Bronconeumol. 2011;47:539–40.
Tomaszek SC, Huebner M, Wigle DA. Prospects for molecular staging of non-small-cell lung cancer from genomic alterations. Expert Rev Respir Med. 2010;4:499–508.
Chatterjee M, Sequeira L, Jenkins-Kabaila M, Dubyk CW, Pathak S, van Golen KL. Individual rac GTPases mediate aspects of prostate cancer cell and bone marrow endothelial cell interactions. J Signal Transduct. 2011;2011:541851.
Hwang SL, Chang JH, Cheng TS, Sy WD, Lieu AS, Lin CL, Lee KS, Howng SL, Hong YR. Expression of Rac3 in human brain tumors. J Clin Neurosci. 2005;12:571–4.
Zhang G, Shang B, Yang P, Cao Z, Pan Y, Zhou Q. Induced pluripotent stem cell consensus genes: implication for the risk of tumorigenesis and cancers in induced pluripotent stem cell therapy. Stem Cells Dev. 2012;21:955–64.
Tjandra H, Compton J, Kellogg D. Control of mitotic events by the Cdc42 GTPase, the Clb2 cyclin and a member of the PAK kinase family. Curr Biol. 1998;8:991–1000.
Zhu WL, Hossain MS, Guo DY, Liu S, Tong H, Khakpoor A, Casey PJ, Wang M. A role for Rac3 GTPase in the regulation of autophagy. J Biol Chem. 2011;286:35291–8.
Fernández Larrosa PN, Ruiz Grecco M, Alvarado CV, Micenmacher S, Aguirre C, Martínez Noel G, Costas MA, Rubio MF. Rapamycin effect on senescence and autophagy processes in human cell lines. Medicina (B Aires). 2011;71:238–42.
Acknowledgments
This work was supported by the National Natural Science Foundation of China (Grant No. 30700821), the Liaoning Bai Qian Wan Talents Program (Grant No. 2011921038), the Liaoning Province Science and Technology (S&T) Project (Grant No. 2013225585), and the Liaoning Province Natural Science Foundation (Grant No. 2015020464).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of interest
None.
Rights and permissions
About this article
Cite this article
Wang, G., Wang, H., Zhang, C. et al. Rac3 regulates cell proliferation through cell cycle pathway and predicts prognosis in lung adenocarcinoma. Tumor Biol. 37, 12597–12607 (2016). https://doi.org/10.1007/s13277-016-5126-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-016-5126-7