Abstract
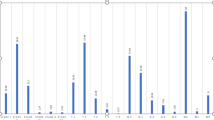
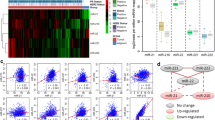
This study aimed to screen potential microRNAs (miRNAs) and genes related to human primary breast cancer. The gene and miRNA expression profile data of GSE19783 was obtained from Gene Expression Omnibus. The matched messenger RNA (mRNA) and miRNA expression profiles of 100 human primary breast cancer samples were chosen for further analysis. The miRNA-gene regulatory modules were screened via iterative multiplicative updating algorithm. The potential functions of genes in modules were predicted by functional and pathway enrichment analysis; meanwhile, the potential functions of miRNAs were predicted by functional enrichment analysis. Furthermore, miRNA-miRNA functional synergistic network and miRNA-miRNA co-regulatory network were constructed. Totally, 16 miRNA-gene modules were screened, containing 222 miRNA-gene interactions. The genes in these modules were mainly related to breast cancer. Genes in module 6 (e.g., SFRP1) were enriched in cell junction assembly; genes in module 8 and 12 (e.g., ESR1 and ERBB4) were significantly implicated in mammary gland alveolus and lobule development. Meanwhile, genes in module 12 (e.g., ERBB4) were enriched in the pathway of endocytosis. Besides, several miRNAs (e.g., miR-375) were enriched in inflammatory cell apoptotic process; some other miRNAs (e.g., miR-139-5p and miR-9) were enriched in response to vitamin D. Additionally, miR-139-5p with several other miRNAs (e.g., miR-9) co-regulated SFRP1; miR-375, miR-592, and miR-135a co-regulated ESR1 and ERBB4. Some miRNAs (e.g., miR-139-5p and miR-9) and their target gene SFRP1, as well as several other miRNAs (e.g., miR-375, miR-592, and miR-135a) and their target genes (e.g., ESR1 and ERBB4), might be crucial in the pathogenesis of primary breast cancer.


Similar content being viewed by others
References
Siegel R, Ma J, Zou Z, Jemal A. Cancer statistics, 2014. CA Cancer J Clin. 2014;64(1):9–29.
Sieuwerts AM, Mostert B, Bolt-de Vries J, Peeters D, de Jongh FE, Stouthard JM, et al. mRNA and microRNA expression profiles in circulating tumor cells and primary tumors of metastatic breast cancer patients. Clin Cancer Res. 2011;17(11):3600–18.
Muller A, Homey B, Soto H, Ge N, Catron D, Buchanan ME, et al. Involvement of chemokine receptors in breast cancer metastasis. Nature. 2001;410(6824):50–6.
Zhang Z, Ni C, Chen W, Wu P, Wang Z, Yin J, et al. Expression of CXCR4 and breast cancer prognosis: a systematic review and meta-analysis. BMC Cancer. 2014;14(49):1471–2407.
Hassan S, Buchanan M, Jahan K, Aguilar-Mahecha A, Gaboury L, Muller WJ, et al. CXCR4 peptide antagonist inhibits primary breast tumor growth, metastasis and enhances the efficacy of anti‐VEGF treatment or docetaxel in a transgenic mouse model. Int J Cancer. 2011;129(1):225–32.
Niikura N, Liu J, Hayashi N, Mittendorf EA, Gong Y, Palla SL, et al. Loss of human epidermal growth factor receptor 2 (HER2) expression in metastatic sites of HER2-overexpressing primary breast tumors. J Clin Oncol. 2012;30(6):593–9.
Kasimir-Bauer S, Hoffmann O, Wallwiener D, Kimmig R, Fehm T. Expression of stem cell and epithelial-mesenchymal transition markers in primary breast cancer patients with circulating tumor cells. Breast Cancer Res. 2012;14(1):R15.
Sanz-Pamplona R, Aragüés R, Driouch K, Martín B, Oliva B, Gil M, et al. Expression of endoplasmic reticulum stress proteins is a candidate marker of brain metastasis in both ErbB-2+ and ErbB-2− primary breast tumors. Am J Pathol. 2011;179(2):564–79.
Gonzalez-Angulo AM, Ferrer-Lozano J, Stemke-Hale K, Sahin A, Liu S, Barrera JA, et al. PI3K pathway mutations and PTEN levels in primary and metastatic breast cancer. Mol Cancer Ther. 2011;10(6):1093–101.
Santini D, Schiavon G, Vincenzi B, Gaeta L, Pantano F, Russo A, et al. Receptor activator of NF-kB (RANK) expression in primary tumors associates with bone metastasis occurrence in breast cancer patients. PLoS One. 2011;6(4):e19234.
Iorio MV, Ferracin M, Liu C-G, Veronese A, Spizzo R, Sabbioni S, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65(16):7065–70.
Lehmann U, Streichert T, Otto B, Albat C, Hasemeier B, Christgen H, et al. Identification of differentially expressed microRNAs in human male breast cancer. BMC Cancer. 2010;10(1):109.
Jiang S, Zhang H-W, Lu M-H, He X-H, Li Y, Gu H, et al. MicroRNA-155 functions as an OncomiR in breast cancer by targeting the suppressor of cytokine signaling 1 gene. Cancer Res. 2010;70(8):3119–27.
Enerly E, Steinfeld I, Kleivi K, Leivonen SK, Aure MR, Russnes HG, et al. miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One. 2011;6(2):e16915.
Zhang S, Li Q, Liu J, Zhou XJ. A novel computational framework for simultaneous integration of multiple types of genomic data to identify microRNA-gene regulatory modules. Bioinformatics. 2011;27(13):i401–9.
Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–64.
Smyth GK. Linear models and empirical bayes methods for assessing differential expression in microarray experiments. Stat Appl Genet Mol Biol. 2004;3(1). doi:10.2202/1544-6115.1027.
Xiao F, Zuo Z, Cai G, Kang S, Gao X, Li T. miRecords: an integrated resource for microRNA–target interactions. Nucleic Acids Res. 2009;37:D105–10.
Dweep H, Sticht C, Pandey P, Gretz N. miRWalk–database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44(5):839–47.
Franceschini A, Szklarczyk D, Frankild S, Kuhn M, Simonovic M, Roth A, et al. STRING v9. 1: protein-protein interaction networks, with increased coverage and integration. Nucleic Acids Res. 2013;41:D808–15.
Berry MW, Browne M, Langville AN, Pauca VP, Plemmons RJ. Algorithms and applications for approximate nonnegative matrix factorization. Comput Stat Data Anal. 2007;52(1):155–73.
Davis AP, Murphy CG, Johnson R, Lay JM, Lennon-Hopkins K, Saraceni-Richards C et al. The comparative toxicogenomics database: update 2013. Nucleic Acids Res. 2013;41:D1104–14.
Matys V, Fricke E, Geffers R, Gößling E, Haubrock M, Hehl R, et al. TRANSFAC®: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31(1):374–8.
Chen JS, Hung WS, Chan HH, Tsai SJ, Sun HS. In silico identification of oncogenic potential of fyn-related kinase in hepatocellular carcinoma. Bioinformatics. 2013;29(4):420–7.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. Gene Ontology: tool for the unification of biology. Nat Genet. 2000;25(1):25–9.
Altermann E, Klaenhammer TR. PathwayVoyager: pathway mapping using the Kyoto Encyclopedia of Genes and Genomes (KEGG) database. BMC Genomics. 2005;6(1):60.
Kohl M, Wiese S, Warscheid B. Cytoscape: software for visualization and analysis of biological networks. In: Hamacher M, Eisenacher M, Stephan C, editors. Data Mining in Proteomics, vol 696. New York: Springer; 2011. p. 291–303.
Joshi PA, Khokha R. The mammary stem cell conundrum: is it unipotent or multipotent? Breast Cancer Res. 2012;14(2):305.
Veeck J, Niederacher D, An H, Klopocki E, Wiesmann F, Betz B, et al. Aberrant methylation of the Wnt antagonist SFRP1 in breast cancer is associated with unfavourable prognosis. Oncogene. 2006;25(24):3479–88.
Huth L, Rose M, Kloubert V, Winkens W, Schlensog M, Hartmann A, et al. BDNF is associated with SFRP1 expression in luminal and basal-like breast cancer cell lines and primary breast cancer tissues: a novel role in tumor suppression? PLoS One. 2014;9(7):e102558.
Hoevel T, Macek R, Swisshelm K, Kubbies M. Reexpression of the TJ protein CLDN1 induces apoptosis in breast tumor spheroids. Int J Cancer. 2004;108(3):374–83.
Sauer T, Pedersen M, Ebeltoft K, Naess O. Reduced expression of Claudin‐7 in fine needle aspirates from breast carcinomas correlate with grading and metastatic disease. Cytopathology. 2005;16(4):193–8.
Colston K, Berger U, Coombes RC. Possible role for vitamin D in controlling breast cancer cell proliferation. Lancet. 1989;333(8631):188–91.
Dalessandri KM, Miike R, Wiencke JK, Farren G, Pugh TW, Manjeshwar S, et al. Vitamin D receptor polymorphisms and breast cancer risk in a high-incidence population: a pilot study. J Am Coll Surg. 2012;215(5):652–7.
Krishnan K, Steptoe AL, Martin HC, Pattabiraman DR, Nones K, Waddell N, et al. miR-139-5p is a regulator of metastatic pathways in breast cancer. RNA. 2013;19(12):1767–80.
Zhou X, Marian C, Makambi KH, Kosti O, Kallakury BV, Loffredo CA, et al. MicroRNA-9 as potential biomarker for breast cancer local recurrence and tumor estrogen receptor status. PLoS One. 2012;7(6):e39011.
Croce CM. Oncogenes and cancer. N Engl J Med. 2008;358(5):502–11.
Holst F, Stahl PR, Ruiz C, Hellwinkel O, Jehan Z, Wendland M, et al. Estrogen receptor alpha (ESR1) gene amplification is frequent in breast cancer. Nat Genet. 2007;39(5):655–60.
Shao J, Li S, Crowder R, Kitchens R, Johnson S, Goncalves R et al. Abstract S3-05: Patient-derived xenograft study reveals endocrine therapy resistance of ER+ breast cancer caused by distinct ESR1 gene aberrations. Cancer Res. 2013;73(24 Supplement):S3-05-S3-.
Guler G, Iliopoulos D, Guler N, Himmetoglu C, Hayran M, Huebner K. Wwox and Ap2γ expression levels predict tamoxifen response. Clin Cancer Res. 2007;13(20):6115–21.
Kurppa KJ, Rokavec M, Sundvall M, Kellokumpu-Lehtinen P-L, Joensuu H, Brauch H, et al. ERBB4 promoter polymorphism is associated with poor distant disease-free survival in high-risk early breast cancer. PLoS One. 2014;9(7):e102388.
Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140(6):883–99.
de Souza Rocha Simonini P, Breiling A, Gupta N, Malekpour M, Youns M, Omranipour R, et al. Epigenetically deregulated microRNA-375 is involved in a positive feedback loop with estrogen receptor alpha in breast cancer cells. Cancer Res. 2010;70(22):9175–84.
Romero-Cordoba S, Rodriguez-Cuevas S, Rebollar-Vega R, Quintanar-Jurado V, Maffuz-Aziz A, Jimenez-Sanchez G, et al. Identification and pathway analysis of microRNAs with no previous involvement in breast cancer. PLoS One. 2012;7(3):e31904.
Chen Y, Zhang J, Wang H, Zhao J, Xu C, Du Y, et al. miRNA-135a promotes breast cancer cell migration and invasion by targeting HOXA10. BMC Cancer. 2012;12(1):111.
Acknowledgments
This study was supported by Hubei Provincial Natural Science Foundation of China (No. 2014CFB366).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Yang, Y., Xing, Y., Liang, C. et al. Crucial microRNAs and genes of human primary breast cancer explored by microRNA-mRNA integrated analysis. Tumor Biol. 36, 5571–5579 (2015). https://doi.org/10.1007/s13277-015-3227-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-015-3227-3