Abstract
There is evidence that insertion of viral DNA into a mammalian genome can lead to alterations of methylation patterns. The aim of the present study was to examine the presence of DNA sequences of five human DNA viruses (assessed by PCR): JC polyoma virus (JCV), human adenovirus (AdV), Epstein–Barr virus (EBV), Kaposi sarcoma-associated herpesvirus (KSHV/HHV8) and human papillomavirus (HPV) in a cohort of 186 sporadic colorectal cancers (CRCs) and related these data with the methylation status of six CpG island methylator phenotype (CIMP)-specific genes (MLH1, CACNA1G, NEUROG1, IGF2, SOCS1, RUNX3) and seven cancer-related genes markers (p16, MINT1, MINT2, MINT31, EN1, SCTR and INHBB) assessed by methylation-specific PCR in 186 and 134 CRC cases, respectively. The AdV, KSHV and HPV were detected in four (2%), two (1%) and zero CRC cases, respectively, and thus were excluded from further analyses. Although 19% and 9% of the CRCs were positive for EBV and JCV, respectively, no associations between virus presence and CpG island methylation were found after correction for multiple testing. Our results demonstrate that the presence of DNA sequences of JCV and EBV in CRC is unrelated to the methylation of the 13 cancer-related CpG islands and CIMP.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Background
The process of DNA methylation in human cells is controlled by DNA methyltransferases (DNMTs). DNMTs catalyze the transfer of a methyl group from the methyl donor to the 5′ position on the cytosine ring [1]. CpG island methylation is prevalent in human sporadic cancers resulting in the transcriptional silencing of many important genes [2]. Abberant CpG island methylation is one of the hallmarks of sporadic colorectal cancer (CRC), and in this context, it has been observed that a subset of CRCs exhibit an exceptionally high frequency of methylation of discrete CpG islands referred to as the CpG island methylator phenotype (CIMP) [3]. Although the mechanism responsible for abberant CpG island methylation and/or CIMP has not yet been elucidated, there is mounting evidence that infectious agents, such as DNA viruses, may cause aberrant methylation in CRCs [4].
Evidence that DNA viruses influence aberrant methylation comes from the observation that insertion of adenovirus (AdV) DNA into a mammalian genome can lead to alterations of methylation patterns in cellular DNA [5]. Moreover, two recent studies have reported that Epstein–Barr virus (EBV) transformed lymphoblastoid cell lines demonstrate alterations of methylation patterns when compared to peripheral blood leukocytes [6, 7]. Finally, abberant methylation of p16 tumour suppresor gene has been reported in Kaposi sarcoma-associated herpesvirus (KSHV/HHV8) associated primary efusion lymphoma. It was also demonstrated repeatedly that various viral proteins interact with a number of host epigenetic regulators which may contribute to alternations in DNA methylation and histone modifications [8]. The AdV small E1A protein binds to DNA methyltransferase 1, increases its activity and alters global patterns of histone modification [9]. The human papillomavirus type 16 (HPV16) E6 oncoprotein interacts with and thus inhibits the histone acetyltransferase CBP/p300 complex, whereas the E7 oncoprotein binds to the histone deacetylase complex Mi2β [10]. The KSHV (HHV8) LANA protein activates DNMT3a facilitating de novo promoter methylation [11]. The EBV latent membrane protein 1 activates all three human DNMTs, which increases de novo promoter methylation [12]. Although none of the JC virus (JCV) proteins have been reported to interact with any epigenetic regulator, some previous reports have suggested a link between expression of the JCV T-antigene and extensive promoter methylation leading to methylator phenotype (CIMP) in CRC [13]. In this study, we assess the presence of DNA sequences from five viruses (JCV, EBV, AdV, KSHV (HHV8) and HPV) in CRCs in relation to methylation of a number of cancer-related genes as well as to CIMP.
Methods
Patients
The study was performed on 186 surgically resected frozen tissues of sporadic CRCs that were obtained between 2001 and 2008 from the 2nd Department of General and Oncological Surgery, Wroclaw Medical University and the Department of General Surgery, Regional Specialized Hospital, Wroclaw.
The CRC patient’s group consisted entirely of Polish individuals (all Caucasians). Only patients with primary, sporadic colorectal cancer who had not received preoperative therapy were included into the studies. Informed consent was obtained from all patients. The study was accepted by the Wroclaw Medical University Ethics Committee.
BRAF V600E mutation
Detection of BRAF V600E in tumour tissues was carried out using a procedure originally described by Sapio et al. [14]. Briefly, mutant allele-specific PCR was used to amplify the exon 15 region of the BRAF gene. PCR products with an expected size of 125 bp were resolved on a 2.5% agarose–ethidium bromide gel.
Bisulphite Treatment of DNA, Methylation-Specific PCR and CIMP
Bisulphite treatment of genomic DNA obtained from resected frozen tumour tissues was carried out using a procedure described by Chan et al. [15]. Approximately 50 ng of the modified DNA was amplified in a PTC 200 DNA Engine Thermal cycler (MJ Research, Inc. Waltham, MA, USA) with primers specific to either the methylated or unmethylated promoter sequences of the CACNA1G, IGF2, NEUROG1, RUNX3, SOCS1, hMLH1, p16, MINT1, MINT2, MINT31, EN1, SCTR and INHBB genes. The primer sequences and amplification conditions of methylation-specific PCR utilized in this study are described elsewhere [3, 15, 16]. CIMP was defined by the use of a specific panel of markers and criteria described by Weisenberger et al. [3]. Briefly, after the analysis of the methylation of a panel of five markers (CACNA1G, IGF2, NEUROG1, RUNX3 and SOCS1), CIMP+ tumours were defined as those with at least three methylated CIMP markers. CIMP− tumours were defined as those with at most two methylated CpG islands.
Viral DNA amplification
Firstly, all samples were examined for DNA integrity using amplification of the β-globin gene. Great care was taken to avoid specimen contamination during the course of this investigation. Sample preparation, electrophoresis and PCR solution preparation were conducted in physically separated laboratory areas. The viral sequences were amplified using a single PCR approach to avoid false positives and ensure reliable PCR detection. For JCV, KSHV (HHV8) and EBV, we used species-specific PCR primers [13, 17]. As the HPV and AdV are species with a high degree of genetic heterogeneity, we used broad-spectrum PCR primers that permit the simultaneous amplification of a range of HPV or AdV types in a single PCR test [18, 19]. A negative control was included in each PCR assay. All samples were re-examined for the presence of viral DNA. All positive samples for EBV and JCV were confirmed using restriction enzyme digestion.
All PCR reactions were carried out separately in a 25-μL reaction volume containing 1× PCR buffer (Qiagen), 1.5 mmol/L MgCl2, 200 μmol/L deoxynucleoside triphosphate, 50 ng of genomic DNA, 0.2 μmol/L of each primer and 0.75 U of Hot-Start DNA polymerase (Qiagen). All PCR reactions were run under the conditions described in the references, except for an initial denaturation period of 15 min at 95°C, which we applied in all tests. Primer sequences and related references are shown in Table 1. All positive controls were purchased from Advanced Biotechnologies Inc (Columbia, MD, USA) except of HPV, where DNA obtained from larynx tumour positive for HPV 16 was applied (see Table 1).
Statistical analysis
The Pearson chi-squared test (if all expected cell frequencies were ≥5) or Fisher’s exact test was used to test whether the presence of a virus is associated with the methylation of a CpG island or CIMP status. All p values were two-sided and Bonferroni correction was taken into consideration because of the multiple comparisons carried out in this study. The R statistical package was used to carry out the necessary statistical tests and calculate the confidence intervals for the odds ratio.
Results
Study group and CIMP classification
We examined the CIMP status of 186 sporadic CRCs by methylation-specific PCR using a CIMP-specific marker panel (CACNA1G, IGF2, NEUROG1, RUNX3 and SOCS1) [3]. The characteristics of study group, both overall and with respect to CIMP status, are shown in Table 2. On the average, the CIMP+ cases tended to be older than CIMP− cases; however, these differences did not attain statistical significance. There were no significant differences between CIMP− and CIMP+ cases in sex distribution. Since CIMP was defined as the presence of at least three methylated sites out of the five studied loci, 25% (46 out of 186) of the tumours were classified as CIMP+. A strongly bimodal distribution of tumours according to the number of methylated loci was observed (Fig. 1). To examine whether our CIMP classification was adequate, we determined the presence of the BRAF V600E mutation and MLH1 methylation. We observed a significant association of both the BRAF V600E mutation and MLH1 methylation with CIMP+ tumours (odds ratio (OR) = 12.33, 95% confidence interval (CI) = 4.21–41.54, P = 1.8 × 10−7 and OR = 15.5, 95% CI = 4.54–68.65, P = 2.8 × 10−7, respectively). These results, together with the strongly bimodal distribution of the number of methylated tumours in our cohort, proved that our CIMP classification was appropriate.
Detection of viral DNA sequences in colorectal cancers
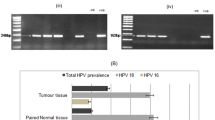
In the current study, we analysed 186 cases of sporadic colorectal tumours (all amplifiable with a β-globin primer set) for the presence of five viruses (JCV, AdV, EBV, KSHV (HHV8) and HPV) using PCR. The AdV, KSHV and HPV were detected in four (2%), two (1%) and zero CRC cases, respectively, and thus, these species were excluded from the statistical analyses. Nineteen percent (36 out of 186) cases were positive for EBV, and 9% (17 out of 186) of the cases were positive for JCV. There was no relationship between the presence of EBV and JCV and sex (see Table 3).
Relationship of JCV and EBV with CIMP and methylation in CIMP markers
The overall results on the presence of EBV and JCV according to CIMP and methylation of CIMP-specific markers are presented in Table 3 and Fig. 1. The presence of JCV was significantly less common in tumours with at least one methylated CIMP-specific marker (5.6% (7 out of 124); P = 0.03) than in the group without methylated CIMP-specific markers (16% (10 out of 62); Fig. 1). We also examined the relationship of EBV and JCV with methylation for each CIMP-specific marker individually (Table 3). In this analysis, EBV was inversely associated with methylation of RUNX3 (P = 0.048), and JCV was inversely associated with methylation of CACNA1G (P = 0.03). However, these associations became nonsignificant after correction for multiple testing.
Relationship of JCV and EBV with methylation in individual CpG islands
Because JCV and EBV have been implicated in CpG island methylation, we examined the relationship between the presence of these two species and methylation in seven individual CpG islands (Table 4). For this analysis, we were able to use 134 of the 186 CRCs due to the limited availability of DNA from the tumours. In this analysis, JCV showed an inverse association with methylation of MINT1 (P = 0.04). However, this association became nonsignificant after correction for multiple testing
Conclusions
Epigenetic alternations play a key role in the process of colorectal carcinogenesis [20]. The recently described CIMP in sporadic CRC is an alternative tumorigenesis pathway characterized by the methylation of multiple promoter regions of tumour suppressor genes harbouring CpG islands [3]. Paradoxically, despite dozens of studies, the defects in the methylation machinery leading to CIMP in CRC remain still unidentified [4].
Several lines of evidence suggest that a number of viral proteins interact with the host’s epigenetic machinery, most likely to evade detection by the host’s immune system [8]. Moreover, experimental approaches, such as transfection of mammalian cells with an adenovirus, demonstrate that insertion of viral DNA can alter the host’s DNA methylation patterns regardless of the transcription state of the viral genome [5, 21]. Further evidence of virus-induced alternations in host’s DNA methylation patterns was observed in EBV transformed lymphoblastoid cells [6, 7]. Therefore, the DNA viruses such as JCV, AdV, EBV, KSHV (HHV8) and HPV seemed to be reasonable candidates for being a causative agent of alterations in DNA methylation resulting in CIMP in sporadic CRC.
In the present study, AdV and KSHV (HHV8) were detected in four and two CRC cases, respectively. Hence, these species were excluded from further analyses. Of note, the very low frequency of infections in our CRC group by these species are in agreement with those reported by Knösel et al. [22], who studied various infectious pathogens in Crohn’s disease.
In our CRC group, no positive signals (0 out of 186) were obtained for HPV. HPV DNA has been detected in CRCs by others with a frequency ranging from 50% to 80% [23–25]. Interestingly, in the all of above-mentioned reports, a nested PCR was employed to detect HPV. In our opinion, this detection technique is likely to introduce a high rate of false positives or the detection of insignificant virus load in the sample, which leads to overestimation of virus presence in the sample; therefore, we decided to stay in line with a single PCR approach for all virus species investigated in this study. In agreement with our strategy, the results of Atula et al. [26] suggest that HPV DNA detected by nested PCR in laryngeal carcinoma cell lines is likely to relate to the presence of minimal amounts of HPV (20 HPV-positive cells among 106 tumour cells) suggesting non-clonal persistence of HPV in laryngeal carcinomas. Most importantly, we were able to amplify HPV DNA from larynx cancer samples by our PCR approach (data not shown) which further suggests lack or minimal amounts of HPV copies in our CRC cohort.
Of the other viruses examined, EBV was the most frequent pathogen (19% of the cases). Few papers have been also published on the presence of EBV in CRCs, moreover with contradicting results. EBV DNA has been detected in CRCs with a frequency ranging from 0% to 19% [27–29]. In great majority of above-mentioned studies, EBV has been detected by using in situ hybridisation of the small EBV-encoded RNA1 (EBER1) which is believed to be consistently expressed in EBV infection. However, an EBER-negative form of EBV infection has been observed in breast cancers and hepatocellular carcinoma, and therefore, a lack of detectable EBNA1 expression cannot be taken as proof of absence of the virus [30, 31]. Since in the present study we used DNA obtained from resected tumours, it is possible that some of the EBV-positive signals reflect the presence of tumour-infiltrating lymphocytes (TILs) in the tumour stroma that carry EBV rather than the presence of EBV-positive tumour cells. However, Yoshiyama et al. [32] and others reported that EBV is able to infect epithelial cell lines in vitro only by co-culture with EBV-infected B cells; therefore, the contribution of EBV-positive TILs (as possible EBV transmitters) in pathogenesis of colon cancer cannot be completely excluded [33]. In general, EBV was not associated either with the CIMP phenotype or methylation of any of the individual CpG islands studied, except for an inverse association with methylation in one of the CIMP-specific markers (RUNX3; nonsignificant after correction for multiple testing).
The second most frequent virus in our CRC group was JCV (9% of the cases). As in the case of prevalence of HPV in CRCs, the papers published on the presence of JCV T-antigen sequence in CRCs display contradictory results with frequencies ranging from 0% in Italian and Spanish study, through 26% in Japanese cases to 77% in American patients [13, 34–36]. Given the all above-cited studies based on relatively similar PCR approaches, the discrepancies of JCV frequencies in CRC may reflect ethnic-dependent epidemiology of JCV or lack of reliable and reproducible test for the detection of JCV DNA [37]. A previous study by Goel et al. [13] reported an association between JCV T-antigen expression and methylation of the promoter region of various cancer-related genes in colorectal cancer. Goel et al. [13] also hypothesized that the JCV T-antigen may be responsible for induction of the methylator phenotype in CRC. Contrary to these results, we found that the presence of JCV T-antigen sequence was less common in tumours with at least one methylated CIMP-specific marker. Moreover, we showed that among the CRC cases, the presence of JCV T-antigen sequence was inversely associated with methylation in two CIMP-related genes (CACNA1G and MINT1). Although these associations became nonsignificant after correction for multiple testing, it did not escape our attention that the presence of JCV T-antigen sequence shows some tendency towards unmethylated CpG islands. Similar results were presented in a very recent paper by Nosho et al. [38], who observed an inverse association of JCV T-antigen expression with CIMP and a lack of association with methylation in 16 genes in a large sample of CRCs. Interestingly, some recent reports show very strong evidence that JCV T-antigen interacts with p53 and Rb family proteins and therefore may induce chromosomal instability (CIN), which is a largely independent from the CIMP pathway in colorectal carcinogenesis [39, 40]. The independence of the CIN and CIMP pathways in sporadic CRC manifests itself via the high level of chromosomal aberrations in tumours with none of the CIMP markers methylated [41]. The more common presence of JCV in a group of CRCs without any CIMP marker methylated revealed by our study provides some support to the notion that JCV may contribute to CIN in a fraction of CRC cases [38].
A potential limitation of this study is that DNA methylation investigation was limited to several CpG islands; therefore, the influence of presence of JCV and EBV on DNA methylation in CRC needs to be further elucidated on the genome-wide scale. Finally, further research is needed to assess the relationship between virus load and DNA methylation in CRC.
In summary, our study provides no evidence of involvement of AdV, KSHV (HHV8) and HPV in pathogenesis of CRC in Polish population. The presence of JCV and EBV sequences in CRCs was not related to methylation of the 13 cancer-related CpG islands and CIMP.
References
Bestor TH. The DNA methyltransferases of mammals. Hum Mol Genet. 2000;9:2395–402.
Karpinski P, Sasiadek MM, Blin N. Aberrant epigenetic patterns in the etiology of gastrointestinal cancers. J Appl Genet. 2008;49:1–10.
Weisenberger DJ, Siegmund KD, Campan M, et al. CpG island methylator phenotype underlies sporadic microsatellite instability and is tightly associated with BRAF mutation in colorectal cancer. Nat Genet. 2006;38:787–93.
Issa JP. CpG island methylator phenotype in cancer. Nat Rev Cancer. 2004;4:988–93.
Doerfler W. Epigenetic mechanisms in human adenovirus type 12 oncogenesis. Semin Cancer Biol. 2009;19:136–43.
Brennan EP, Ehrich M, Brazil DP, et al. Comparative analysis of DNA methylation profiles in peripheral blood leukocytes versus lymphoblastoid cell lines. Epigenetics. 2009;3:159–64.
Grafodatskaya D, Choufani S, Ferreira JC, Butcher DT, Lou Y, Zhao C, et al. EBV transformation and cell culturing destabilizes DNA methylation in human lymphoblastoid cell lines. Genomics. 2010;2:73–83.
Flanagan JM. Host epigenetic modifications by oncogenic viruses. Br J Cancer. 2007;29:183–8.
Horwitz GA, Zhang K, McBrian MA, Grunstein M, Kurdistani SK, Berk AJ. Adenovirus small e1a alters global patterns of histone modification. Science. 2008;22:1084–5.
Zimmermann H, Degenkolbe R, Bernard HU, O’Connor MJ. The human papillomavirus type 16 E6 oncoprotein can down-regulate p53 activity by targeting the transcriptional coactivator CBP/p300. J Virol. 1999;73:6209–19.
Shamay M, Krithivas A, Zhang J, Hayward SD. Recruitment of the de novo DNA methyltransferase Dnmt3a by Kaposi’s sarcoma-associated herpesvirus LANA. PNAS. 2006;26:14554–9.
Tsai CL, Li HP, Lu YJ, et al. Activation of DNA methyltransferase 1 by EBV LMP1 involves c-Jun NH2-terminal kinase signaling. Cancer Res. 2006;15:11668–76.
Goel A, Li MS, Nagasaka T, Shin SK, Fuerst F, Ricciardiello L, et al. Association of JC virus T-antigen expression with the methylator phenotype in sporadic colorectal cancers. Gastroenterology. 2006;130:1950–61.
Sapio MR, Posca D, Troncone G, Pettinato G, Palombini L, Rossi G, et al. Detection of BRAF mutation in thyroid papillary carcinomas by mutant allele-specific PCR amplification (MASA). Eur J Endocrinol. 2006;154:341–8.
Chan A, Issa JP, Morris JS, Hamilton SR, Rashid A. Concordant CpG island methylation in hyperplastic polyposis. Am J Pathol. 2002;160:529–36.
Karpinski P, Ramsey D, Grzebieniak Z, Sasiadek MM, Blin N. The CpG island methylator phenotype correlates with long-range epigenetic silencing in colorectal cancer. Mol Cancer Res. 2008;6:585–91.
Pan L, Milligan L, Michaeli J, Cesarman E, Knowles DM. Polymerase chain reaction detection of Kaposi’s sarcoma-associated herpesvirus-optimized protocols and their application to myeloma. J Mol Diagn. 2001;3:32–8.
Fujinaga Y, Shimada M, Okazawa K, Fukushima M, Kato I, Fujinaga K. Simultaneous detection and typing of genital human papillomavirus DNA using the polymerase chain reaction. J Gen Virol. 1991;72:1039–44.
Echavarria M, Forman M, Ticehurst J, Dumler JS, Charache P. PCR method for detection of adenovirus in urine of healthy and human immunodeficiency virus-infected individuals. J Clin Microbiol. 1998;36:3323–6.
Schuebel KE, Chen W, Cope L, et al. Comparing the DNA hypermethylome with gene mutations in human colorectal cancer. PLoS Genet. 2007;3:1709–23.
Müller K, Heller H, Doerfler W. Foreign DNA integration. Genome-wide perturbations of methylation and transcription in the recipient genomes. J Biol Chem. 2001;27:14271–8.
Knösel T, Schewe C, Petersen N, Dietel M, Petersen I. Prevalence of infectious pathogens in Crohn’s disease. Pathol Res Pract. 2009;205:223–30.
Cheng JY, Sheu LF, Meng CL, Lee WH, Lin JC. Detection of human papillomavirus DNA in colorectal carcinomas by polymerase chain reaction. Gut. 1995;37:87–90.
Bodaghi S, Yamanegi K, Xiao SY, Da Costa M, Palefsky JM, Zheng ZM. Colorectal papillomavirus infection in patients with colorectal cancer. Clin Cancer Res. 2005;11:2862–7.
Damin DC, Caetano MB, Rosito MA, Schwartsmann G, Damin AS, Frazzon AP, et al. Evidence for an association of human papillomavirus infection and colorectal cancer. Eur J Surg Oncol. 2007;33:569–74.
Atula S, Grenman R, Kujari H, Syrjänen S. Detection of human papillomavirus (HPV) in laryngeal carcinoma cell lines provides evidence for a heterogeneic cell population. Eur J Cancer. 1999;5:825–32.
Cho YJ, Chang MS, Park SH, Kim HS, Kim WH. In situ hybridization of Epstein–Barr virus in tumor cells and tumor-infiltrating lymphocytes of the gastrointestinal tract. Hum Pathol. 2001;32:297–301.
Kijima Y, Hokita S, Takao S, et al. Epstein–Barr virus involvement is mainly restricted to lymphoepithelial type of gastric carcinoma among various epithelial neoplasms. J Med Virol. 2001;4:513–8.
Liu HX, Ding YQ, Li X, Yao KT. Investigation of Epstein–Barr virus in Chinese colorectal tumors. World J Gastroenterol. 2003;9:2464–8.
Bonnet M, Guinebretiere J-M, Kremmer E, Grunewald V, Benhamou E, Contesso G, et al. Detection of Epstein–Barr virus in invasive breast cancers. J Natl Cancer Inst. 1999;91:1376–81.
Sugawara Y, Mizugaki Y, Uchida T, Torii T, Imai S, Makuuchi M, et al. Detection of Epstein–Barr virus (EBV) in hepatocellular carcinoma tissue: a novel EBV latency characterized by the absence of EBV-encoded small RNA expression. Virology. 1999;256:196–202.
Yoshiyama H, Imai S, Shimizu N, Takada K. Epstein–Barr virus infection of human gastric carcinoma cells: implication of the existence of a new virus receptor different from CD21. J Virol. 1997;7:5688–91.
Shannon-Lowe CD, Neuhierl B, Baldwin G, Rickinson AB, Delecluse HJ. Resting B cells as a transfer vehicle for Epstein–Barr virus infection of epithelial cells. PNAS. 2006;18:7065–70.
Hernández Losa J, Fernandez-Soria V, Parada C, Sanchez-Prieto R, Ramón Y, Cajal S, et al. JC virus and human colon carcinoma: an intriguing and inconclusive association. Gastroenterology. 2003;1:268–9.
Hori R, Murai Y, Tsuneyama K, et al. Detection of JC virus DNA sequences in colorectal cancers in Japan. Virchows Arch. 2005;4:723–30.
Giuliani L, Ronci C, Bonifacio D, Di Bonito L, Favalli C, Perno CF, et al. Detection of oncogenic DNA viruses in colorectal cancer. Anticancer Res. 2008;28(2B):1405–10.
Burnett-Hartman AN, Newcomb PA, Potter JD. Infectious agents and colorectal cancer: a review of Helicobacter pylori, Streptococcus bovis, JC virus, and human papillomavirus. Cancer Epidemiol Biomarkers Prev. 2008;17:2970–9.
Nosho K, Shima K, Kure S, et al. JC virus T-antigen in colorectal cancer is associated with p53 expression and chromosomal instability, independent of CpG island methylator phenotype. Neoplasia. 2009;11:87–95.
Ricciardiello L, Baglioni M, Giovannini C, et al. Induction of chromosomal instability in colonic cells by the human polyomavirus JC virus. Cancer Res. 2003;63:7256–62.
Del Valle L, Khalili K. Detection of human polyomavirus proteins, T-antigen and agnoprotein, in human tumor tissue arrays. J Med Virol. 2010;5:806–11.
Cheng YW, Pincas H, Bacolod MD, et al. CpG island methylator phenotype associates with low-degree chromosomal abnormalities in colorectal cancer. Clin Cancer Res. 2008;14:6005–13.
Acknowledgements
The authors would like to acknowledge the contribution of Prof. Zygmunt Grzebieniak and Dr. Tadeusz Sebzda to the data collection. This study was supported by grants from the State Committee for Scientific Research, Polish Ministry for Scientific Research and Information Technology (No 1423/P01/2007/32, 2007–2010 and 6041/B/P01/2010/38 2010–2013).
Conflicts of interest
None
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Karpinski, P., Myszka, A., Ramsey, D. et al. Detection of viral DNA sequences in sporadic colorectal cancers in relation to CpG island methylation and methylator phenotype. Tumor Biol. 32, 653–659 (2011). https://doi.org/10.1007/s13277-011-0165-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13277-011-0165-6