Abstract
Background
Heparan sulfate 2-O-sulfotransferase 1 (HS2ST1) catalyzes the sulfation of glucuronic acid residues in heparan sulfate proteoglycans, enabling these proteoglycans to interact with numerous ligands within tumor microenvironments. However, the prognostic role of HS2ST1 expression in cancer remains unclear.
Objective
This investigated HS2ST1 expression levels and their prognostic significance in various cancer types, demonstrated the prognostic value of HS2ST1 expression in hepatocellular carcinoma (HCC) patients, and identified molecular signatures associated with HS2ST1 expression.
Methods
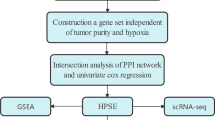
HS2ST1 expression and patient survival data from The Cancer Genome Atlas (TCGA) datasets were analyzed using the Gene Expression Profiling Interactive Analysis (GEPIA) portal. We obtained gene expression and clinicopathological information on HCC patients from the TCGA and the Japan and France International Cancer Genome Consortium (ICGC) databases and performed survival analyses. We also examined relevant protein networks, differentially expressed genes, gene set enrichments, and tumor immune microenvironment features associated with HS2ST1 expression.
Results
HS2ST1 exhibited higher expression in eight tumor types compared with normal tissues and was associated with poor prognoses in five tumors, including HCC. HS2ST1 status correlated with poor prognosis in two ICGC HCC cohorts. Elevated HS2ST1 expression in HCC tumors was associated with signaling pathways involved in cell cycle progression, protein secretion, and mTORC1 signaling. Moreover, HS2ST1 expression levels were inversely correlated with immune cell infiltration in the tumor microenvironment.
Conclusion
Our study elucidates the prognostic significance of HS2ST1 expression in HCC patients and provides insights into the potential roles of HS2ST1 in signaling pathways and the tumor microenvironment.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
The tumor microenvironment comprises a heterogeneous population of proliferating tumor cells, infiltrating immune cells, and fibroblasts; extracellular matrix (ECM); a basement membrane; vasculature; and secreted factors (Sarrazin et al. 2011). Heparan sulfate proteoglycans (HSPGs) are essential for ECM assembly and signaling in such microenvironments. HSPGs are glycoproteins consisting of core proteins covalently attached to heparan sulfate chains, thus categorizing them as glycosaminoglycans. The 13 core HSPG proteins are distributed across various cellular compartments, including cell and basement membranes, the ECM, and secretory vesicles. HSPGs bind cytokines, chemokines, and growth factors; the bound materials serve as reservoirs from which regulatory factors can be released via selective degradation of the heparan sulfate chains. HSPGs also function as receptors for proteases and protease inhibitors. Membrane HSPGs, in combination with integrin and adhesion molecules, facilitate cell-ECM and cell–cell interactions; they serve both as co-receptors of various growth factor receptors and as endocytic receptors that clear bound ligands.
Most interactions between HSPGs and protein ligands occur through the heparan sulfate domains of HSPGs, although some ligands bind directly to the core proteins. The biosynthesis of heparan sulfate chains begins with posttranslational modifications in the Golgi apparatus upon delivery of the core proteins from the endoplasmic reticulum (De Pasquale and Pavone 2020). Tetrasaccharide linkers and the first N-acetyl-d-glucosamine residues are attached to the serine residues of core proteins. Subsequently, d-glucuronic acid and N-acetyl-d-glucosamine residues are alternately added to growing chains. Concurrently, each chain undergoes sequential modifications that include deacetylation, epimerization of d-glucuronic acid to l-iduronic acid residues, and sulfation. N-deacetylase/N-sulfotransferase 1–4 (NDST1-4) mediates the addition of sulfate to the free amino groups of N-acetyl-d-glucosamine residues from which acetyl groups have been removed. Heparan sulfate 2-O-sulfotransferase 1 (HS2ST1) catalyzes 2-O-sulfation at iduronic acid and, less frequently, glucuronic acid residues in heparan sulfate chain. HS6ST1-3 and HS3ST1-6 facilitate 6-O and 3-O sulfation at glucosamine residues, respectively. These modified domains appear in clusters of varying lengths, interspersed with unaltered segments; the domains create binding sites for protein ligands such as antithrombin, fibroblast growth factor (FGF), and the FGF receptor.
Uronyl 2-O sulfotransferase 1 (also known as HS2ST1) is the only enzyme that catalyzes the 2-O sulfation of iduronic acid residues in heparan sulfate chains. The perinatal lethality of Hs2st1-deficient mice is characterized by bilateral renal agenesis and defects of the eyes and skeleton (Kuehn et al. 2021). Liver-specific alterations in mouse Hs2st1 levels affect lipoprotein clearance (Anower et al. 2019). However, the role of HS2ST1 in cancer remains poorly understood and appears to vary by cancer type. HS2ST1 exhibited higher expression in osteosarcoma compared with normal tissue, but lower tumor expression levels correlated with poor prognosis in high-risk patients (Huang et al. 2022; Yang et al. 2022). HS2ST1 expression was associated with favorable prognoses in multiple myeloma patients (Bret et al. 2009). In contrast, breast cancer patients with high HS2ST1 expression had a worse prognosis than those with lower HS2ST1 expression (Kuehn et al. 2021; Teixeira et al. 2020; Kumar et al. 2020). Because the sulfation pattern is typically determined by the cell type in which heparan sulfate is expressed (Sarrazin et al. 2011), it is crucial to study the clinical significance of HS2ST1 expression in relation to the specific tumor of interest.
In this study, we investigated HS2ST1 expression and the prognostic value of HS2ST1 expression across The Cancer Genome Atlas (TCGA) cohorts. We discovered that HS2ST1 expression correlated with poor prognosis in the TCGA-Liver Hepatocellular Carcinoma (LIHC) cohort and was prognostically relevant in two independent International Cancer Genome Consortium (ICGC) cohorts of hepatocellular carcinoma (HCC) patients. Further analyses of the protein–protein networks and gene sets involved, as well as immune cell infiltration, provided insights into the biological processes affected by HS2ST1 expression.
Materials and methods
Data collection and preprocessing
TCGA datasets from the Gene Expression Profiling Interactive Analysis (GEPIA) portal (http://gepia2.cancer-pku.cn/#index) were used to analyze differences in HS2ST1 expression levels between normal and tumor tissues, and patient prognoses across different cancers (Tang et al. 2019). Gene expression and clinical data from the TCGA-LIHC and ICGC databases of Japan and France (ICGC-LIRI-JP and ICGC-LICA-FR, respectively) were also collected and analyzed. The TCGA sample data were acquired using the UCSC Xena Browser (http://xena.ucsc.edu/) (Goldman et al. 2020). After removal of samples lacking survival data, information on 365 primary solid tumor samples was used in subsequent analyses. The ICGC cohort data were downloaded (https://dcc.icgc.org/) (Zhang et al. 2019) and curated using the same approach. Data from 231 Japanese and 98 French primary tumor samples were subjected to further analyses. RNA-seq data were preprocessed and normalized using DESeq2 software (Love et al. 2014).
Survival analysis
Patients were stratified into HS2ST1-high- and -low-expression groups based on cutoffs determined using the maximally selected rank statistics of the Survminer package (version 0.4.9). This method yields an optimal cutoff point for evaluating overall survival. Kaplan–Meier (KM) analysis was performed and log-rank tests were conducted to evaluate the associations between different HS2ST1 expression levels and overall survival in HCC patients.
Protein network, differentially expressed gene (DEG), and gene set enrichment analyses
Protein–protein interaction analysis utilized the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING) database (version 12) (Szklarczyk et al. 2023) to identify interactions between HS2ST1 and other proteins. This analysis provided insights into the roles and potential protein partners of HS2ST1 in molecular pathways, enhancing the overall understanding of HS2ST1 biological functions and their relevance to various cellular processes.
To identify HCC biological pathways involving HS2ST1, DEGs between the high- and low-HS2ST1 expression groups of the TCGA cohort were identified using DESeq2 software (Love et al. 2014); the criteria were an adjusted P-value < 0.05 and an absolute log2-fold change of 1. Gene set enrichment analysis (GSEA) software version 4.3.2 was used to compare the high‐ and low‐HS2ST1-expression groups (Mootha et al. 2003; Subramanian et al. 2005). The hallmark gene set of the Molecular Signatures Database was utilized in the analysis (Liberzon et al. 2011, 2015; Subramanian et al. 2005).
Links between tumor immunity and HS2ST1 expression
The Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) algorithm (Yoshihara et al. 2013) was used to explore the relationship between HS2ST1 expression levels, the tumor microenvironment, and the extent of tumor immune cell infiltration. Subsequently, the Cell-type Identification By Estimating Relative Subsets Of RNA Transcripts (CIBERSORT) algorithm (Newman et al. 2015) was applied to determine the relative scores of 22 different immune cells in the TCGA-LIHC cohort. Additionally, the TISIDB database (Ru et al. 2019), an integrated repository of tumor-immune system interactions, was utilized to investigate the relationship between immunomodulator levels and HS2ST1 mRNA expression levels.
Statistical analysis
All statistical analyses were conducted within R (version 4.4.0) and RStudio (version 2024.04.0) softwares. Survival analyses were performed using the R packages ‘Survival’ (version 3.5–8) and ‘Survminer’ (version 0.4.9). The significance threshold for all tests was set to P < 0.05.
Results
HS2ST1 expression levels in various cancers
HS2ST1 expression levels in tumors were compared with those in adjacent normal tissues of the TCGA datasets and normal tissues of the GTEx dataset across various tissue types in the GEPIA2 portal. HS2ST1 expression levels differed among cancer types (Fig. 1). Of the 31 cancers studied, HS2ST1 expression levels in tumor tissues were higher (compared with normal tissue) in patients with diffuse large B-cell lymphoma (DLBC), esophageal carcinoma (ESCA), glioblastoma multiforme (GBM), pancreatic adenocarcinoma (PAAD), rectal adenocarcinoma (READ), skin cutaneous melanoma (SKCM), stomach adenocarcinoma (STAD), and thymoma (THYM). However, the reverse was observed in patients with acute myeloid leukemia (LAML) and pheochromocytoma and paraganglioma (PCPG).
HS2ST1 expression levels in tumor and normal tissues. HS2ST1 expression levels [log2(TPM + 1) values] in tumor tissues and adjacent normal tissues of the TCGA datasets and normal tissues of the GTEx dataset (31 different tissue types) were analyzed using the GEPIA2 portal. Only cancers exhibiting significant differences (*: P < 0.05) in HS2ST1 expression between tumor and normal tissues are shown. DLBC diffuse large B-cell lymphoma, ESCA esophageal carcinoma, GBM glioblastoma multiforme, PAAD pancreatic adenocarcinoma READ rectal adenocarcinoma, SKCM skin cutaneous melanoma, STAD stomach adenocarcinoma, THYM thymoma, LAML acute myeloid leukemia, PCPG pheochromocytoma and paraganglioma, num(T) number of TCGA tumor samples, num(N) number of normal TCGA and GTEx samples
Prognostic value of HS2ST1 expression in TCGA cohorts
We examined whether HS2ST1 expression was associated with prognosis in various cancers. Survival analysis of GEPIA2 data, using median values as cutoffs, revealed that HS2ST1 expression levels were prognostic in six cancer types (Fig. 2). Patients with high HS2ST1 expression in kidney chromophobe (KICH), lower-grade brain glioma (LGG), LIHC, sarcoma (SARC), and uveal melanoma (UVM) had poor prognoses. High HS2ST1 expression was associated with favorable prognosis only in patients with kidney renal clear cell carcinoma (KIRC). Thus, HS2ST1 expression correlated with poor prognosis in patients with certain tumors. Because LIHC is a highly malignant tumor associated with high mortality, we also validated the prognostic role of HS2ST1 expression in HCC.
Prognostic impact of HS2ST1 expression in TCGA cancer types. KM analysis and log-rank tests were conducted using the medians as the GEPIA2 cutoff points. Only cancers exhibiting significant differences (*P < 0.05) are shown. a KICH, kidney renal clear cell carcinoma, b LGG, brain lower grade glioma, c LIHC liver hepatocellular carcinoma; d SARC sarcoma, e UVM uveal melanoma, f KIRC kidney renal clear cell carcinoma. HR hazard ratio
Prognostic value of HS2ST1 status in various HCC cohorts
We explored whether HS2ST1 mRNA expression levels predicted the prognosis of HCC patients in the TCGA-LIHC, ICGC-LIRI-JP, and ICGC-LICA-FR cohorts (Table 1). We utilized optimal cutoffs of HS2ST1 expression levels identified by maximally selected rank statistics to maximize statistical power and flexibility afforded by the various datasets. In the TCGA-LIHC cohort, 69 primary tumor samples exhibited high HS2ST1 expression and 296 exhibited low expression. Survival analysis revealed a significant association between HS2ST1 expression levels and overall survival (P = 0.0023) (Fig. 3a). Lower HS2ST1 expression was protective; higher expression correlated with shorter survival of HCC patients. In the LIRI-JP and LICA-FR cohorts, higher HS2ST1 expression correlated significantly with poorer overall survival (P = 0.00033 and P = 0.0023, respectively) (Figs. 3b and c). Thus, HS2ST1 expression levels were prognostically significant across different HCC cohorts and might aid patient stratification.
Protein–protein interaction, DEG, and gene set enrichment analyses
To explore whether HS2ST1 expression levels affected prognosis, we used STRING database to investigate protein–protein interactions involving HS2ST1 and compared DEGs of patients exhibiting high and low HS2ST1 expression. HS2ST1 interacts with various enzymes that both biosynthesize and modify HSPGs, including exostosin glycosyltransferase 1 (EX1), glucuronic acid epimerase (GLCE), N-deacetylase/N-sulfotransferase (NDST), heparan sulfate-glucosamine 3-sulfotransferase (HS3ST), and heparan sulfate 6-O-sulfotransferase (HS6ST) (Fig. 4a). These proteins are known to interact with HS2ST1 but provide limited information in interpreting the prognostic role of HS2ST1 in HCC.
Functional analysis of the protein network, DEGs, and enriched gene sets associated with HS2ST1 expression. a Protein–protein interaction analysis using the STRING database revealed a network of proteins associated with HS2ST1. b Volcano plot highlighting DEGs between the high- and low-HS2ST1 expression groups with adjusted P values < 0.05 and |log2-fold changes (LFCs)| ≥ 1. c Functional GSEA comparing the high- and low-HS2ST1 expression groups; highlighted pathways were enriched in the high-HS2ST1 expression group. FDR false discovery rate, NES normalized enrichment score
To further explore how high HS2ST1 expression levels might contribute to the poor prognosis of HCC patients, we identified DEGs in the TCGA-LIHC cohort and enriched gene sets in the high-HS2ST1 expression group. DEG analysis revealed that genes encoding chromogranin A (CHGA), GRM7 glutamate metabotropic receptor 7 (GRM7), CPA2 carboxypeptidase A2 (CPA2), calcitonin receptor (CALCR), and solute carrier family 5 member 12 (SLC5A12) were more highly expressed. Conversely, genes encoding ADCY8 adenylate cyclase 8 (ADCY8), kallikrein-related peptidase 4 (KTP4), lipocalin 2 (LCN2), aldehyde dehydrogenase 3 family member A1 (ALDH3A1), atypical chemokine receptor 1 (ACKR1), and solute carrier family 4 member 3 (SLC4A3) exhibited lower expression in the high-HS2ST1 expression group than in the low-HS2ST1 expression group (Fig. 4b).
Next, GSEA was performed using the hallmark gene set (Fig. 4c). The G2M checkpoint, E2F target, protein secretion, and mTORC1 signaling pathways were significantly enriched in the high-HS2ST1 expression group compared with the low-HS2ST1 expression group across all three cohorts. The activities of the unfolded protein response and glycolysis pathways were significantly enhanced in the LIRI-JP cohort exhibiting high HS2ST1 expression compared with the low-HS2ST1 expression group.
Immune cell infiltration and HS2ST1 expression levels
Hs2st1 facilitates immune cell infiltration in mouse models of airway inflammation (Axelsson et al. 2012; Ge et al. 2018). Therefore, we explored whether HS2ST1 status affected the tumor microenvironment. The stromal (P = 0.0022) and immune scores (P = 0.0272) of the ESTIMATE algorithm were lower in the high-HS2ST1 expression group. The ESTIMATE score, which combines stromal and immune scores to derive a measure of tumor purity, indicated fewer nontumor cells in the high-HS2ST1 expression group (P = 0.0068) (Fig. 5a).
Association of the tumor immune microenvironment with HS2ST1 expression in the TCGA-LIHC cohort. a Stromal, immune, and ESTIMATE scores determined by the ESTIMATE algorithm for the high- and low-HS2ST1 expression groups (*P < 0.05). b Infiltration levels of various immune cell types according to HS2ST1 expression status, as revealed by the CIBERSORT algorithm (*P < 0.05). c Correlations between the expression of HS2ST1 and immune checkpoint proteins, as revealed by the TISIDB (*P < 0.05)
We used CIBERSORT software to identify specific immune cell populations associated with HS2ST1 expression (Fig. 5b). The high-HS2ST1 expression group contained fewer activated mast cells but more plasma B cells and resting mast cells compared with the low-HS2ST1 expression group. Next, we used the TISIDB database to reveal correlations between HS2ST1 expression levels and those of key immune checkpoint proteins (Fig. 5c). HS2ST1 expression levels were weakly negatively correlated with those of PD-1 (PDCD1) (rho = − 0.172, P = 0.000852) and CTLA-4 (rho = − 0.102, P = 0.049). The correlation between HS2ST1 expression and that of CD274 (also known as PD-L1) was marginally nonsignificant (rho = 0.099, P = 0.0552). These findings underscore the complex interplay between HS2ST1 expression levels and the HCC microenvironment.
Discussion
Sulfation of HSPG heparan chains critically mediates interactions among growth factors, cytokines, and immune cells. The roles of HS2ST1 in cancer and the overall immune landscape remain poorly understood. In this study, we used TCGA datasets to identify tumors in which HS2ST1 was highly expressed or HS2ST1 levels were prognostic. The role of HS2ST1 status as a negative prognostic factor in TCGA-LIHC was validated in two ICGC HCC patient cohorts. High HS2ST1 expression in HCC tumors was associated with cell signaling that enhanced cell proliferation and protein secretion, as well as changes in immune cell components in the tumor microenvironment.
Glypican-3 expression levels are higher in HCC than in surrounding normal tissues; thus, glypican-3 is a HSPG commonly targeted by biologics such as chimeric antigen receptors, bispecific antibodies, and immunotoxins (Fu et al. 2023). HS2ST1 expression was higher in tumor tissues compared with normal tissues in patients with diffuse large B-cell lymphomas, esophageal carcinomas, glioblastoma multiforme, pancreatic adenocarcinomas, rectal adenocarcinomas, skin cutaneous melanomas, stomach adenocarcinomas, and thymomas. Normal tissues adjacent to diffuse large B-cell lymphomas and thymomas expressed very low levels of HS2ST1, suggesting that HS2ST1 can serve as a cancer-specific marker in such patients. Higher HS2ST1 expression was associated with poor prognoses in patients with kidney chromophobe syndrome, lower-grade gliomas, HCCs, sarcomas, and uveal melanomas. To extend the reliability of this evidence across cohorts of different nationalities, we evaluated the prognostic value of HS2ST1 expression levels in HCC cohorts from Japan and France. Higher HS2ST1 levels robustly predicted poorer survival.
Liver HSPGs interact with many growth factors (e.g., basic FGF, hepatocyte growth factors, platelet-derived growth factors, vascular endothelial growth factors), transforming growth factor-β and cytokines (CCL5 and CXCL12) (Lai et al. 2004, 2008). Such HSPG interactions, along with those involving growth factor receptors, facilitate growth factor retention, protect factors from degradation, and ultimately trigger tumor suppression. For example, HS2ST1 overexpression decreased FGF-2/receptor interaction (Kumar et al. 2020), in turn reducing FGFR-MAPK signaling and invasion by MDA-MB-231 and MCF7 cells. Both HS2ST1 and syndecan-1 promoted FGFR1 endocytosis and inhibited FGFR1-Akt signaling in MCF7 cells (Kang et al. 2020). Interactions between heparan sulfate chains and proteases facilitate allosteric protease activation that promotes tumor progression. Syndecan-2 enhances pro-MMP-7 processing and the subsequent cleavage of the E-cadherin substrate, enhancing colon cancer cell migration (Sarrazin et al. 2011).
Currently, the binding specificities between sulfate groups in the heparan sulfate chains of HSPGs and ligands are not fully understood in the HCC context. Increased sulfation associated with decreased expression of sulfatase-1, an extracellular enzyme that remodels HSPGs, enhanced FGF- and HGF-mediated signaling, and increased HCC cell growth (Lai et al. 2004). Conversely, enhanced sulfation caused by sulfatase-2 knockdown inhibited HCC cell proliferation and migration via FGF2-ERK signaling (Lai et al. 2008). Therefore, sulfation-regulating enzymes may selectively affect ligand binding to HSPGs. Our GSEA results revealed that increased HS2ST1 expression activated mTORC1 signaling, E2F-dependent gene expression, G2M checkpoint and protein secretion in three cohorts of HCC patients. Additionally, we identified DEGs associated with high HS2ST1 expression, including several types of proteases (kallikrein-related peptidase 4 and carboxypeptidase A2), receptors (atypical chemokine receptor 1 and the calcitonin receptor), and signaling molecules (adenylate cyclase 8, chromogranin A, and lipocalin 2). HS2ST1 may regulate the expression of these proteins and thus affect HCC tumor progression. These results suggest that HS2ST1-mediated 2-O-sulfuration of iduronic acid residues in HCC patients promotes growth factor signaling, cell proliferation, and protein secretion. However, the DEGs identified in the TCGA-LIHC cohort were not found in the other ICGC cohorts, suggesting further studies are warranted to identify plausible partners of HS2ST1 regardless of tumor heterogeneity.
We found that tumors expressing high levels of HS2ST1 exhibited limited stromal cell infiltration and less infiltration of activated mast cells compared with those with low HS2ST1 expression (Fig. 5). Such suppression of the immune environment may explain the poor prognostic value of HS2ST1 status in HCC patients. Previous studies using mouse airway inflammation models also supported an immunosuppressive role for Hs2st1. Inactivation of mouse endothelial cell Hs2st1 triggered elevated neutrophil trafficking via L-selectin-dependent rolling of neutrophils and increased binding of IL-8 and macrophage inflammatory protein-2 to endothelial cells (Axelsson et al. 2012). Leukocyte and endothelial cell Hs2st1 deficiencies in a mouse model of allergic asthma enhanced eosinophil recruitment associated with persistent inflammation (Ge et al. 2018). Further work is needed to elucidate the interactions between immune cells and 2-O-sulfated uronic acid residues in heparan sulfate chains and the associations between immune checkpoint proteins and HS2ST1 in the tumor microenvironment.
Considering the therapeutic benefits provided by an understanding of how heparin pentasaccharide binds to antithrombin, further exploration of HS2ST1/ligand interactions could prove highly informative. Although our single-gene analysis provided accurate assessments of prognostic risks in independent, large patient cohorts, it remains unclear whether alterations in HS2ST1 expression directly influence disease progression or if other underlying genetic factors are involved. To address these uncertainties and efficiently elucidate the specific biological pathways involved, multi-gene analysis of the interconnected HSPG network is necessary. Our study elucidates the prognostic value of HS2ST1 status in patients with specific tumors, including HCC. HS2ST1 expression was associated with increased signaling related to E2F-mediated gene expression, protein secretion, and mTORC1 signaling, as well as reduced immune cell infiltration. These results enhance our understanding of HS2ST1's roles in HCC and the tumor microenvironment, providing a rationale for therapeutic applications of HS2ST1.
Data availability
The datasets presented in this study can be found in online repositories. TCGA and GTEx datasets are available on GEPIA2. The RNASeq and clinical data for TCGA-LIHC are available on UCSC Xena Browser (http://xena.ucsc.edu/). The RNASeq and clinical data for ICGC-LIRI-JP and ICGC-LICA-FR were downloaded from the ICGC data portal (https://dcc.icgc.org/). All other data supporting the findings of this study are available within the article.
References
Anower-E-Khuda F, Singh G, Deng Y, Gordts PLSM, Esko JD (2019) Triglyceride-rich lipoprotein binding and uptake by heparan sulfate proteoglycan receptors in a CRISPR/Cas9 library of Hep3B mutants. Glycobiology 29:582–592. https://doi.org/10.1093/glycob/cwz037
Axelsson J, Xu D, Kang BN, Nussbacher JK, Handel TM, Ley K, Sriramarao P, Esko JD (2012) Inactivation of heparan sulfate 2-O-sulfotransferase accentuates neutrophil infiltration during acute inflammation in mice. Blood 120:1742–1751. https://doi.org/10.1182/blood-2012-03-417139
Bret C, Hose D, Reme T, Sprynski AC, Mahtouk K, Schved JF, Quittet P, Rossi JF, Goldschmidt H, Klein B (2009) Expression of genes encoding for proteins involved in heparan sulphate and chondroitin sulphate chain synthesis and modification in normal and malignant plasma cells. Br J Hematol 145:350–368. https://doi.org/10.1111/j.1365-2141.2009.07633.x
De Pasquale V, Pavone LM (2020) Heparan sulfate proteoglycan signaling in tumor microenvironment. Int J Mol Sci 21:6588. https://doi.org/10.3390/ijms21186588
Fu Q, Zheng Y, Fang W, Zhao Q, Zhao P, Liu L, Zhai Y, Tong Z, Zhang H, Lin M et al (2023) RUNX-3-expressing CAR T cells targeting glypican-3 in patients with heavily pretreated advanced hepatocellular carcinoma: a phase I trial. EClinicalMedicine 63:102175. https://doi.org/10.1016/j.eclinm.2023.102175
Ge XN, Bastan I, Ha SG, Greenberg YG, Esko JD, Rao SP, Sriramarao P (2018) Regulation of eosinophil recruitment and allergic airway inflammation by heparan sulfate proteoglycan (HSPG) modifying enzymes. Exp Lung Res 44:98–112. https://doi.org/10.1080/01902148.2018.1451574
Goldman MJ, Craft B, Hastie M, Repecka K, McDade F, Kamath A, Banerjee A, Luo Y, Rogers D, Brooks AN et al (2020) Visualizing and interpreting cancer genomics data via the Xena platform. Nat Biotechnol 38:675–678. https://doi.org/10.1038/s41587-020-0546-8
Huang W, Xiao Y, Wang H, Chen G, Li K (2022) Identification of risk model based on glycolysis-related genes in the metastasis of osteosarcoma. Front Endocrinol (lausanne) 13:1047433. https://doi.org/10.3389/fendo.2022.1047433
Kang D, Jung SH, Lee GH, Lee S, Park HJ, Ko YG, Kim YN, Lee JS (2020) Sulfated syndecan 1 is critical to preventing cellular senescence by modulating fibroblast growth factor receptor endocytosis. FASEB J 34:10316–10328. https://doi.org/10.1096/fj.2019027
Kuehn J, Espinoza-Sanchez NA, Teixeira F, Pavao MSG, Kiesel L, Gyorffy B, Greve B, Gotte M (2021) Prognostic significance of hedgehog signaling network-related gene expression in breast cancer patients. J Cell Biochem 122:577–597. https://doi.org/10.1002/jcb.29886
Lai JP, Chien JR, Moser DR, Staub JK, Aderca I, Montoya DP, Matthews TA, Nagorney DM, Cunningham JM, Smith DI et al (2004) hSulf1 Sulfatase promotes apoptosis of hepatocellular cancer cells by decreasing heparin-binding growth factor signaling. Gastroenterology 126:231–248. https://doi.org/10.1053/j.gastro.2003.09.043
Lai JP, Sandhu DS, Yu C, Han T, Moser CD, Jackson KK, Guerrero RB, Aderca I, Isomoto H, Garrity-Park MM et al (2008) Sulfatase 2 upregulates glypican 3, promotes fibroblast growth factor signaling, and decreases survival in hepatocellular carcinoma. Hepatology 47:1211–1222. https://doi.org/10.1002/hep.22202
Liberzon A, Subramanian A, Pinchback R, Thorvaldsdottir H, Tamayo P, Mesirov JP (2011) Molecular signatures database (MSigDB) 3.0. Bioinformatics 27:1739–1740. https://doi.org/10.1093/bioinformatics/btr260
Liberzon A, Birger C, Thorvaldsdottir H, Ghandi M, Mesirov JP, Tamayo P (2015) The molecular signatures database (MSigDB) hallmark gene set collection. Cell Syst 1:417–425. https://doi.org/10.1093/bioinformatics/btr260
Love MI, Huber W, Anders S (2014) Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. https://doi.org/10.1186/s13059-014-0550-8
Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, Puigserver P, Carlsson E, Ridderstrale M, Laurila E et al (2003) PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet 34:267–273. https://doi.org/10.1038/ng1180
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12:453–457. https://doi.org/10.1038/nmeth.3337
Ru B, Wong CN, Tong Y, Zhong JY, Zhong SSW, Wu WC, Chu KC, Wong CY, Lau CY, Chen I et al (2019) TISIDB: an integrated repository portal for tumor-immune system interactions. Bioinformatics 35:4200–4202. https://doi.org/10.1093/bioinformatics/btz210
Sarrazin S, Lamanna WC, Esko JD (2011) Heparan sulfate proteoglycans. Cold Spring Harb Perspect Biol 3:a004952. https://doi.org/10.1101/cshperspect.a004952
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550. https://doi.org/10.1073/pnas.0506580102
Szklarczyk D, Kirsch R, Koutrouli M, Nastou K, Mehryary F, Hachilif R, Gable AL, Fang T, Doncheva NT, Pyysalo S et al (2023) The STRING database in 2023: protein-protein association networks and functional enrichment analyses for any sequenced genome of interest. Nucleic Acids Res 51:D638–D646. https://doi.org/10.1093/nar/gkac1000
Tang Z, Kang B, Li C, Chen T, Zhang Z (2019) GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 47:W556–W560. https://doi.org/10.1093/nar/gkz430
Teixeira F, Vijaya Kumar A, Kumar Katakam S, Cocola C, Pelucchi P, Graf M, Kiesel L, Reinbold R, Pavao MSG, Greve B et al (2020) The Heparan sulfate sulfotransferases HS2ST1 and HS3ST2 are novel regulators of breast cancer stem-cell properties. Front Cell Dev Biol 8:559554. https://doi.org/10.3389/fcell.2020.559554
Vijaya Kumar A, Brezillon S, Untereiner V, Sockalingum GD, Kumar Katakam S, Mohamed HT, Kemper B, Greve B, Mohr B, Ibrahim SA et al (2020) HS2ST1-dependent signaling pathways determine breast cancer cell viability, matrix interactions, and invasive behavior. Cancer Sci 111:2907–2922. https://doi.org/10.1111/cas.14539
Yang G, Jiang J, Yin R, Li Z, Li L, Gao F, Liu C, Zhan X (2022) Two novel predictive biomarkers for osteosarcoma and glycolysis pathways: a profiling study on HS2ST1 and SDC3. Medicine (baltimore) 101:e30192. https://doi.org/10.1097/MD.0000000000030192
Yoshihara K, Shahmoradgoli M, Martinez E, Vegesna R, Kim H, Torres-Garcia W, Trevino V, Shen H, Laird PW, Levine DA et al (2013) Inferring tumor purity and stromal and immune cell admixture from expression data. Nat Commun 4:2612. https://doi.org/10.1038/ncomms3612
Zhang J, Bajari R, Andric D, Gerthoffert F, Lepsa A, Nahal-Bose H, Stein LD, Ferretti V (2019) The international cancer genome consortium data portal. Nat Biotechnol 37:367–369. https://doi.org/10.1038/s41587-019-0055-9
Funding
Not applicable.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
None.
Ethical approval
Not applicable.
Informed consent
All authors consent to the publication of this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chung, T.T., Kim, S.K. & Lee, S.J. Prognostic significance of HS2ST1 expression in patients with hepatocellular carcinoma. Genes Genom (2024). https://doi.org/10.1007/s13258-024-01556-0
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13258-024-01556-0