Abstract
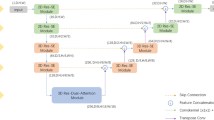
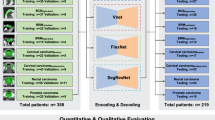
This study aimed to investigate the robustness of a deep learning (DL) fusion model for low training-to-test ratio (TTR) datasets in the segmentation of gross tumor volumes (GTVs) in three-dimensional planning computed tomography (CT) images for lung cancer stereotactic body radiotherapy (SBRT). A total of 192 patients with lung cancer (solid tumor, 118; part-solid tumor, 53; ground-glass opacity, 21) who underwent SBRT were included in this study. Regions of interest in the GTVs were cropped based on GTV centroids from planning CT images. Three DL models, 3D U-Net, V-Net, and dense V-Net, were trained to segment the GTV regions. Nine fusion models were constructed with logical AND, logical OR, and voting of the two or three outputs of the three DL models. TTR was defined as the ratio of the number of cases in a training dataset to that in a test dataset. The Dice similarity coefficients (DSCs) and Hausdorff distance (HD) of the 12 models were assessed with TTRs of 1.00 (training data: validation data: test data = 40:20:40), 0.791 (35:20:45), 0.531 (31:10:59), 0.291 (20:10:70), and 0.116 (10:5:85). The voting fusion model achieved the highest DSCs of 0.829 to 0.798 for all TTRs among the 12 models, whereas the other models showed DSCs of 0.818 to 0.804 for a TTR of 1.00 and 0.788 to 0.742 for a TTR of 0.116, and an HD of 5.40 ± 3.00 to 6.07 ± 3.26 mm better than any single DL models. The findings suggest that the proposed voting fusion model is a robust approach for low TTR datasets in segmenting GTVs in planning CT images of lung cancer SBRT.








Similar content being viewed by others
References
Sethi T (2002) Lung cancer • introduction. Thorax 57:992–993. https://doi.org/10.1136/thorax.57.11.992
Sung H, Ferlay J, Siegel RL et al (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71:209–249
American Cancer Society (2022) Cancer facts & Figs. 2022. American Cancer Society, Atlanta
Detterbeck FC, Mazzone PJ, Naidich DP et al (2013) Screening for lung cancer: diagnosis and management of lung cancer: American college of chest Physicians evidence-based clinical practice guidelines. Chest 143:e78S – e92. https://doi.org/10.1378/chest.12-2350
Crabtree TD, Denlinger CE, Meyers BF et al (2010) Stereotactic body radiation therapy versus surgical resection for stage I non–small cell lung cancer. J Thorac Cardiovasc Surg 140:377–386. https://doi.org/10.1016/j.jtcvs.2009.12.054
Timmerman R, McGarry R, Yiannoutsos C et al (2006) Excessive toxicity when treating central tumors in a phase II study of stereotactic body radiation therapy for medically inoperable early-stage lung cancer. J Clin Oncol 24:4833–4839. https://doi.org/10.1200/jco.2006.07.5937
Weiss E, Hess CF (2003) The impact of gross tumor volume (GTV) and clinical target volume (CTV) definition on the total accuracy in radiotherapy. Strahlenther Onkol 179:21–30. https://doi.org/10.1007/s00066-003-0976-5
Vinod SK, Jameson MG, Min M et al (2016) Uncertainties in volume delineation in radiation oncology: a systematic review and recommendations for future studies. Radiother Oncol 121:169–179. https://doi.org/10.1016/j.radonc.2016.09.009
Van de Steene J, Linthout N, De Mey J et al (2002) Definition of gross tumor volume in lung cancer: inter-observer variability. Radiother Oncol 62:37–49. https://doi.org/10.1016/s0167-8140(01)00453-4
Caldwell CB, Mah K, Ung YC et al (2001) Observer variation in contouring gross tumor volume in patients with poorly defined non-small-cell lung tumors on CT: the impact of 18FDG-hybrid PET fusion. Int J Radiat Oncol Biol Phys 51:923–931. https://doi.org/10.1016/S0360-3016(01)01722-9
Persson GF, Nygaard DE, Hollensen C et al (2012) Interobserver delineation variation in lung tumour stereotactic body radiotherapy. Brit J Radiol 85:e654–e660. https://doi.org/10.1259%2Fbjr%2F76424694
Velazquez ER, Aerts HJWL, Gu Y et al (2012) A semiautomatic CT-based ensemble segmentation of lung tumors: comparison with oncologists’ delineations and with the surgical specimen. Radiother Oncol 105:167–173. https://doi.org/10.1016/j.radonc.2012.09.023
Zhong Z, Kim Y, Plichta K et al (2019) Simultaneous cosegmentation of tumors in PET-CT images using deep fully convolutional networks. Med Phys 46:619–633. https://doi.org/10.1002/mp.13331b
Wong J, Huang V, Giambattista JA et al (2021) Training and validation of deep learning-based auto-segmentation models for lung stereotactic ablative radiotherapy using retrospective radiotherapy planning contours. Front Oncol 11:2085. https://doi.org/10.3389/fonc.2021.626499
Cui Y, Arimura H, Nakano R et al (2021) Automated approach for segmenting gross tumor volumes for lung cancer stereotactic body radiation therapy using CT-based dense V-networks. J Radiat Res 62:346–355. https://doi.org/10.1093/jrr/rraa132
Khorfan R, Kruser TJ, Coughlin JM et al (2020) Survival of primary stereotactic body radiation therapy compared with surgery for operable stage I/II non-small cell lung cancer. Ann Thorac Surg 110(1):228–234. https://doi.org/10.1016/j.athoracsur.2020.01.073
Bardis M, Houshyar R, Chantaduly C et al (2020) Deep learning with limited data: organ segmentation performance by U-Net. Electronics 9:1199. https://doi.org/10.3390/electronics9081199
Narayana PA, Coronado I, Sujit SJ et al (2020) Deep-learning-based neural tissue segmentation of MRI in multiple sclerosis: effect of training set size. J Magn Reson Imaging 51:1487–1496. https://doi.org/10.1002/jmri.26959
Fang Y, Wang J, Ou X et al (2021) The impact of training sample size on deep learning-based organ auto-segmentation for head-and-neck patients. Phys Med Biol. https://doi.org/10.1088/1361-6560/ac2206
Heilemann G, Matthewman M, Kuess P et al (2021) Can generative adversarial networks help to overcome the limited data problem in segmentation? Zeitschrift Med Phys 32:361–368. https://doi.org/10.1016/j.zemedi.2021.11.006
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26:297–302. https://doi.org/10.2307/1932409
Herman GT, Zheng J, Bucholtz CA (1992) Shape-based interpolation. IEEE Comput Graph Appl 12:69–79. https://doi.org/10.1109/38.135915
LeCun Y, Bottou L, Orr GB et al (2002) Efficient backprop. In: Montavon G, Orr G, Müller KR (eds) Neural networks: tricks of the trade, 1st. Springer, Berlin. https://doi.org/10.1007/978-3-642-35289-8_3
Çiçek Ö, Abdulkadir A, Lienkamp SS et al (2016) 3D U-Net: learning dense volumetric segmentation from sparse annotation. Med Image Comput Comput Assist Interv. https://doi.org/10.1007/978-3-319-46723-8_49
Milletari F, Navab N, Ahmadi SA (2016) V-net: fully convolutional neural networks for volumetric medical image segmentation. 3DV. pp 565–571. https://doi.org/10.1109/3DV.2016.79
Gibson E, Giganti F, Hu Y et al (2018) Automatic multi-organ segmentation on abdominal CT with dense V-networks. IEEE Trans Med Imaging 37:1822–1834. https://doi.org/10.1109/TMI.2018.2806309
He K, Zhang X, Ren S et al (2016) Deep residual learning for image recognition. CVPR. https://doi.org/10.1109/CVPR.2016.90
Huang G, Liu Z, Van Der Maaten L et al. (2017) Densely connected convolutional networks. CVPR. pp 4700–4708. https://doi.org/10.1109/CVPR.2017.243
Pedamonti D (2018) Comparison of non-linear activation functions for deep neural networks on MNIST classification task. arXiv. https://doi.org/10.48550/arXiv.1804.02763
Ali R, Ragb HK (2019) Fused deep convolutional neural networks based on voting approach for efficient object classification. NAECON. https://doi.org/10.1109/NAECON46414.2019.9057795
Prechelt L (1998) Early Stopping - But When? Neural Networks: Tricks of the Trade. Lecture Notes in Computer Science. Springer, Berlin. https://doi.org/10.1007/3-540-49430-8_3
Zhu Y, Tan Y, Hua Y et al (2012) Automatic segmentation of ground-glass opacities in lung CT images by using Markov random field-based algorithms. J Digit Imaging 25:409–422. https://doi.org/10.1007/s10278-011-9435-5
Gibson E, Li W, Sudre C et al (2018) NiftyNet: a deep-learning platform for medical imaging. Comput Methods Programs Biomed 158:113–122. https://doi.org/10.1016/j.cmpb.2018.01.025
Lowekamp BC, Chen DT, Ibáñez L et al (2013) The design of SimpleITK. Front Neuroinform 7:45. https://doi.org/10.3389/fninf.2013.00045
Li X, Li B, Tian L et al (2017) Segmentation of ground glass opacity pulmonary nodules based on a modified random walker approach. J Med Imaging Health Inform 7:1589–1593. https://doi.org/10.1166/jmihi.2017.2170
Parker SM, Siochi RA, Wen S et al (2019) Impact of tumor size on local control and pneumonitis after stereotactic body radiation therapy for lung tumors. Pract Radiat Oncol 9:e90–e97. https://doi.org/10.1016/j.prro.2018.09.003
Acknowledgements
This study was partially supported by a grant from the Centre for Clinical and Translational Research of Kyushu University Hospital and JSPS KAKENHI Grant Number 20K08084. The authors are grateful to all members of the Arimura Laboratory (http://web.shs.kyushu-u.ac.jp/~arimura/) and Saga International Heavy Ion Cancer Treatment Foundation for their valuable comments and helpful discussion.
Author information
Authors and Affiliations
Contributions
Conceptualization: HA and YC, data curation: YS, TY, YC and HA, formal analysis: YC and HA, funding acquisition: HA, investigation: YC, HA, methodology: YC and HA, project administration: HA, resources: YC, HA, TY, YS and HY, software: YC, supervision: HA, validation: YC, HA, TY, YS and HY, visualization: YC and HA, writing—original draft: YC and HA.
Corresponding author
Ethics declarations
Competing interests
All authors declare no financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Ethical approval
This study was performed under the approval of an institutional review board of Kyushu University hospital.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Cui, Y., Arimura, H., Yoshitake, T. et al. Deep learning model fusion improves lung tumor segmentation accuracy across variable training-to-test dataset ratios. Phys Eng Sci Med 46, 1271–1285 (2023). https://doi.org/10.1007/s13246-023-01295-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-023-01295-8