Abstract
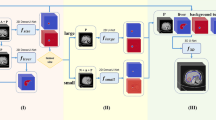
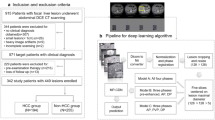
The aim of this study was to develop dual segmentation models for poorly and well-differentiated hepatocellular carcinoma (HCC), using two-step transfer learning (TSTL) based on dynamic contrast-enhanced (DCE) computed tomography (CT) images. From 2013 to 2019, DCE-CT images of 128 patients with 80 poorly differentiated and 48 well-differentiated HCCs were selected at our hospital. In the first transfer learning (TL) step, a pre-trained segmentation model with 192 CT images of lung cancer patients was retrained as a poorly differentiated HCC model. In the second TL step, a well-differentiated HCC model was built from a poorly differentiated HCC model. The average three-dimensional Dice’s similarity coefficient (3D-DSC) and 95th-percentile of the Hausdorff distance (95% HD) were mainly employed to evaluate the segmentation accuracy, based on a nested fourfold cross-validation test. The DSC denotes the degree of regional similarity between the HCC reference regions and the regions estimated using the proposed models. The 95% HD is defined as the 95th-percentile of the maximum measures of how far two subsets of a metric space are from each other. The average 3D-DSC and 95% HD were 0.849 ± 0.078 and 1.98 ± 0.71 mm, respectively, for poorly differentiated HCC regions, and 0.811 ± 0.089 and 2.01 ± 0.84 mm, respectively, for well-differentiated HCC regions. The average 3D-DSC for both regions was 1.2 times superior to that calculated without the TSTL. The proposed model using TSTL from the lung cancer dataset showed the potential to segment poorly and well-differentiated HCC regions on DCE-CT images.









Similar content being viewed by others
References
Sung H, Ferlay J, Siegel R, Laversanne M, Soerjomataram I, Jemal A, Bray F (2021) Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 71(3):209–249. https://doi.org/10.3322/caac.21660
Villanueva A (2019) Hepatocellular carcinoma. N Engl J Med 380:1450–1462. https://doi.org/10.1056/nejmra1713263
World Health Organization Liver Factsheet. Globocan. https://gco.iarc.fr/today/data/factsheets/cancers/11-Liver-fact-sheet.pdf. Accessed Jun 11, 2022.
Kim E, Viatour P (2020) Hepatocellular carcinoma: old friends and new tricks. Exp Mol Med 52:1898–1907. https://doi.org/10.1038/s12276-020-00527-1
Yamamoto M, Yoshida M, Furuse J, Sano K, Ohtsuka M, Yamashita S et al (2021) Clinical practice guidelines for the management of liver metastases from extrahepatic primary cancers 2021. J Hepato-Biliary-Pancreatic Sci 28(1):1–25
The Japan society of hepatology (2017) Clinical practice guidelines for hepatocellular carcinoma 2017. Kanehara & Co. LTD, Tokyo
Tarhan NC, Hatipoğlu T, Ercan E, Bener M, Keleş G, Başaran C et al (2011) Correlation of dynamic multidetector CT findings with pathological grades of hepatocellular carcinoma. Diagn Interv Radiol 17(4):328–333. https://doi.org/10.4261/1305-3825.dir.2682-09.3
Chen W, Zhang T, Xu L, Zhao L, Liu H, Gu LR et al (2021) Radiomics analysis of contrast-enhanced CT for hepatocellular carcinoma grading. Front Oncol 11:660509. https://doi.org/10.3389/fonc.2021.660509
Pérez-Saborido B, de los Galanes SJ, Menéu-Dı́az JC, Romero CJ, Elola-Olaso AM, Suárez YF et al (2007) Tumor recurrence after liver transplantation for hepatocellular carcinoma: recurrence pathway and prognostic factors. Transplant Proc 39(7):2304–2307. https://doi.org/10.1016/j.transproceed.2007.06.059
Shinkawa H, Tanaka S, Kabata D, Takemura S, Amano R, Kimura K et al (2021) The prognostic impact of tumor differentiation on recurrence and survival after resection of hepatocellular carcinoma is dependent on tumor size. Liver cancer 10(5):461–472. https://doi.org/10.1159/000517992
Kim YS, Kim JW, Yoon WS, Kang MK, Lee IK, Kim TH et al (2016) Interobserver variability in gross tumor volume delineation for hepatocellular carcinoma. Strahlenther Onkol 192:714–721. https://doi.org/10.1007/s00066-016-1028-2
Yasaka K, Akai H, Abe O, Kiryu S (2018) Deep learning with CNN showed high diagnostic performance in differentiation of liver masses at dynamic CT. Radiology 286:887–896. https://doi.org/10.1148/radiol.2017170706
Yuan Y (2017) Hierarchical convolutional-deconvolutional neural networks for automatic liver and tumor segmentation. arXiv preprint arXiv:1710.04540.
Li X, Chen H, Qi X, Dou Q, Fu CW, Heng PA (2018) H-Dense UNet: hybrid densely connected UNet for liver and tumor segmentation from CT volumes. IEEE Trans Med Imaging 37(12):2663–2674
Qayyum A, Lalande A, Meriaudeau F (2020) Automatic segmentation of tumors and affected organs in the abdomen using a 3D hybrid model for computed tomography imaging. Comput Biol Med 127:104097. https://doi.org/10.1016/j.compbiomed.2020.104097
Alirr OI (2020) Deep learning and level set approach for liver and tumor segmentation from CT scans. J Appl Clin Med Phys 21(10):200–209. https://doi.org/10.1002/acm2.13003
Bilic P, Christ P F, Vorontsov E, Chlebus G, Chen H, Dou Q et al (2019) The liver tumor segmentation benchmark (LiTS). arXiv preprint arXiv:1901.04056.
Zhang W, Yang R, Liang F, Liu G, Chen A, Wu H et al (2021) Prediction of microvascular invasion in hepatocellular carcinoma with a multi-disciplinary team-like radiomics fusion model on dynamic contrast-enhanced computed tomography. Front Oncol 11:1–11. https://doi.org/10.3389/fonc.2021.660629
Cui Y, Arimura H, Nakano R, Yoshitake T, Shioyama Y, Yabuuchi H (2021) Automated approach for segmenting gross tumor volumes for lung cancer stereotactic body radiation therapy using CT-based dense V-networks. J Radiat Res 62(2):346–355. https://doi.org/10.1093/jrr/rraa132
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26(3):297–302
Rezatofighi H, Tsoi N, Gwak J, Sadeghian A, Reid I, Savarese S (2019) Generalized intersection over union: a metric and a loss for bounding box regression. IEEE/CVF conference on computer vision and pattern recognition (CVPR) Published online 2019. https://doi.org/10.1109/cvpr.2019.00075
Chen X, Sun S, Bai N, Han K, Liu Q, Yao S et al (2021) A deep learning-based auto-segmentation system for organs-at-risk on whole-body computed tomography images for radiation therapy. Radiother Oncol 160:175–184. https://doi.org/10.1016/j.radonc.2021.04.019
Taha AA, Hanbury A (2015) Metrics for evaluating 3D medical image segmentation: analysis, selection, and tool. BMC Med Imaging. https://doi.org/10.1186/s12880-015-0068-x
Dora L, Agrawal S, Panda R, Abraham A (2018) Nested cross-validation based adaptive sparse representation algorithm and its application to pathological brain classification. Expert Syst Appl 114:313–321. https://doi.org/10.1016/j.eswa.2018.07.039
Landis JR, Koch GG (1977) The measurement of observer agreement for categorical data. Biometrics 33(1):159–174
3D Slicer. https://www.slicer.org/
Smith DJ (2018) A nearest-neighbour discretisation of the regularized stokeslet boundary integral equation. J Comput Phys 358:88–102. https://doi.org/10.1016/j.jcp.2017.12.008
Yamada A, Oyama K, Fujita S, Yoshizawa E, Ichinohe F, Komatsu D et al (2019) Dynamic contrast-enhanced computed tomography diagnosis of primary liver cancers using transfer learning of pretrained convolutional neural networks: Is registration of multiphasic images necessary? Int J Comput Assist Radiol Surg 14(8):1295–1301. https://doi.org/10.1007/s11548-019-01987-1
Milletari F, Navab N, Ahmadi SA (2016) V-Net: Fully convolutional neural networks for volumetric medical image segmentation. International conference on 3D vision. IEEE, Stanford, pp 565–571
Tieleman T, Hinton G (2012) Lecture 6.5-RMSProp: divide the gradient by a running average of its recent magnitude. COURSERA Neural Netw Mach Learn 4(2):26–31
Chlap P, Min H, Vandenberg N, Dowling J, Holloway L, Haworth A (2021) A review of medical image data augmentation techniques for deep learning applications. J Med Imaging Radiat Oncol 65:545–563. https://doi.org/10.1111/1754-9485.13261
Gibson E, Li W, Sudre C, Fidon L, Shakir DI, Wang G et al (2018) NiffyNet: a deep-learning platform for medical imaging. Comput Methods Programs Biomed 158:113–122
Rodarmel C, Shan J (2002) Principal component analysis for hyperspectral image classification. Surv L Inf Sci 62(2):115–122
Liu Z, Liu F, Chen W, Liu X, Hou X, Shen J et al (2021) Automatic segmentation of clinical target volumes for post-modified radical mastectomy radiotherapy using convolutional neural networks. Front Oncol 10:581347. https://doi.org/10.3389/fonc.2020.581347
Codella NCF, Gutman D, Celebi ME, Helba B, Marchetti MA et al (2018) Skin lesion analysis toward melanoma detection: a challenge at the 2017 International symposium on biomedical imaging (ISBI), hosted by the international skin imaging collaboration (ISIC). Int Symp Biomed Imagin. https://doi.org/10.1109/ISBI.2018.8363547
Selvaraju RR, Cogswell M, Das A, Vedantam R, Parikh D, Batra D (2020) Grad-CAM: visual explanations from deep networks via gradient-based localization. Int J Comput Vis 128:336–359
Acknowledgements
This study was partially supported by a grant from Center for Clinical and Translational Research of Kyushu University Hospital and JSPS KAKENHI Grant Number JP20K08084. The authors are grateful to all members of the Arimura Laboratory (http://web.shs.kyushu-u.ac.jp/~arimura/ c.jp/~arimura/), Department of radiology, Saga University Hospital for their valuable comments and helpful discussion and to Hirokazu Takahashi, Satoshi Oeda and Hiroshi Isoda, Liver center, Saga University Hospital for their comments for clinical practice of HCC.
Author information
Authors and Affiliations
Contributions
Conceptualization: HA, NN, Data curation: HA, NN, MO, JN, Formal analysis: NN, MO, KO, Funding acquisition: HA, Investigation: NN, HA, Methodology: NN, HA, Project administration: HA, Resources: NN, HA, JN, CY, KN, MO, SK, HI, Software: NN, KN, CY, Supervision: HA, SK, HI, Validation: NN, HA, Visualization: NN, HA, Writing—original draft: NN, HA.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nagami, N., Arimura, H., Nojiri, J. et al. Dual segmentation models for poorly and well-differentiated hepatocellular carcinoma using two-step transfer deep learning on dynamic contrast-enhanced CT images. Phys Eng Sci Med 46, 83–97 (2023). https://doi.org/10.1007/s13246-022-01202-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-022-01202-7