Abstract
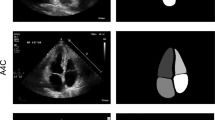
The segmentation of cardiac boundaries, specifically Left Ventricle (LV) segmentation in 2D echocardiographic images, is a critical step in LV segmentation and cardiac function assessment. These images are generally of poor quality and present low contrast, making daily clinical delineation difficult, time-consuming, and often inaccurate. Thus, it is necessary to design an intelligent automatic endocardium segmentation system. The present work aims to examine and assess the performance of some deep learning-based architectures such as U-Net1, U-Net2, LinkNet, Attention U-Net, and TransUNet using the public CAMUS (Cardiac Acquisitions for Multi-structure Ultrasound Segmentation) dataset. The adopted approach emphasizes the advantage of using transfer learning and resorting to pre-trained backbones in the encoder part of a segmentation network for echocardiographic image analysis. The experimental findings indicated that the proposed framework with the \(\text {U}\)-\(\text {Net1}_{\text {VGG19}}\) is quite promising; it outperforms other more recent approaches with a Dice similarity coefficient of 93.30% and a Hausdorff Distance of 4.01 mm. In addition, a good agreement between the clinical indices calculated from the automatic segmentation and those calculated from the ground truth segmentation. For instance, the mean absolute errors for the left ventricular end-diastolic volume, end-systolic volume, and ejection fraction are equal to 7.9 ml, 5.4 ml, and 6.6%, respectively. These results are encouraging and point out additional perspectives for further improvement.







Similar content being viewed by others
Data availability
The dataset used in this study includes 2D echocardiographic images and is publicly accessible. The dataset used in this study can be accessed from: https://camus.creatis.insa-lyon.fr/challenge/.
References
Mc Namara K, Alzubaidi H, Jackson JK (2019) Cardiovascular disease as a leading cause of death: how are pharmacists getting involved? Integr Pharm Res Pract 8:1–11. https://doi.org/10.2147/IPRP.S133088
Sadeghpour A, Alizadehasl A (2022) Practical cardiology. Elsevier, Amsterdam, pp 67–110. https://doi.org/10.1016/B978-0-323-80915-3.00008-9
Carneiro G, Nascimento JC, Freitas A (2012) The segmentation of the left ventricle of the heart from ultrasound data using deep learning architectures and derivative-based search methods. IEEE Trans Image Process 21(3):968–982. https://doi.org/10.1109/TIP.2011.2169273
Dietenbeck T, Alessandrini M, Barbosa D, D’hooge J, Friboulet D, Bernard O (2012) Detection of the whole myocardium in 2d-echocardiography for multiple orientations using a geometrically constrained level-set. Med Image Anal 16(2):386–401. https://doi.org/10.1016/j.media.2011.10.003
Kim T, Hedayat M, Vaitkus VV, Belohlavek M, Krishnamurthy V, Borazjani I (2021) Automatic segmentation of the left ventricle in echocardiographic images using convolutional neural networks. Quant Imaging Med Surg 11(5):1763–1781
Noble JA, Boukerroui D (2006) Ultrasound image segmentation: a survey. IEEE Trans Med Imaging 25(8):987–1010. https://doi.org/10.1109/TMI.2006.877092
Yan J, Zhuang T (2003) Applying improved fast marching method to endocardial boundary detection in echocardiographic images. Pattern Recognit Lett 24(15):2777–2784. https://doi.org/10.1016/S0167-8655(03)00121-1
Paragios N (2003) A level set approach for shape-driven segmentation and tracking of the left ventricle. IEEE Trans Med Imaging 22(6):773–776. https://doi.org/10.1109/TMI.2003.814785
Bosch JG, Mitchell SC, Lelieveldt BPF, Nijland F, Kamp O, Sonka M, Reiber JHC (2002) Automatic segmentation of echocardiographic sequences by active appearance motion models. IEEE Trans Med Imaging 21(11):1374–1383. https://doi.org/10.1109/TMI.2002.806427
Galicia E, Torres F, Escalante B, Olveres J, Arámbula F (2021) Full multi resolution active shape model for left ventricle segmentation. In: Walker A, Rittner L, Romero Castro E, Lepore N, Brieva J, Linguraru MG (eds) 17th international symposium on medical information processing and analysis, p. 41. SPIE, Campinas, Brazil. https://doi.org/10.1117/12.2606252
Ali Y, Beheshti S, Janabi-Sharifi F (2021) Echocardiogram segmentation using active shape model and mean squared eigenvalue error. Biomed Signal Process Control 69:102807. https://doi.org/10.1016/j.bspc.2021.102807
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521(7553):436–444. https://doi.org/10.1038/nature14539
Minaee S, Boykov YY, Porikli F, Plaza AJ, Kehtarnavaz N, Terzopoulos D (2021) Image segmentation using deep learning: a survey. IEEE Trans Pattern Anal Mach Intell. https://doi.org/10.1109/TPAMI.2021.3059968
Smistad E, Ostvik A, Haugen BO, Lovstakken L (2017) 2D left ventricle segmentation using deep learning. In: 2017 IEEE international ultrasonics symposium (IUS), pp. 1–4. IEEE, Washington, DC. https://doi.org/10.1109/ULTSYM.2017.8092573
Leclerc S, Smistad E, Pedrosa J, østvik A, Cervenansky F, Espinosa F, Espeland T, Berg EAR, Jodoin P-M, Grenier T, Lartizien C, D’hooge J, Lovstakken L, Bernard O (2019) Deep learning for segmentation using an open large-scale dataset in 2D echocardiography. IEEE Trans Med Imaging 38(9):2198–2210. https://doi.org/10.1109/TMI.2019.2900516
Dahal L, Kafle A, Khanal B (2020) Uncertainty estimation in deep 2D echocardiography segmentation. arXiv:2005.09349 [cs]. arXiv:2005.09349
Hesse LS, Namburete AIL (2020) Improving U-net segmentation with active contour based label correction. In: Papież BW, Namburete AIL, Yaqub M, Noble JA (eds) Medical image understanding and analysis, vol 1248. Springer, Cham. https://doi.org/10.1007/978-3-030-52791-4_6
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 234–241
Zhao C, Xia B, Chen W, Guo L, Du J, Wang T, Lei B (2021) Multi-scale wavelet network algorithm for pediatric echocardiographic segmentation via hierarchical feature guided fusion. Appl Soft Comput 107:107386. https://doi.org/10.1016/j.asoc.2021.107386
Yang Y, Sermesant M (2021) Shape constraints in deep learning for robust 2D echocardiography analysis. In: Ennis DB, Perotti LE, Wang VY (eds) Functional imaging and modeling of the heart, vol 12738. Springer, Cham, pp 22–34. https://doi.org/10.1007/978-3-030-78710-3_3
Deng K, Meng Y, Gao D, Bridge J, Shen Y, Lip G, Zhao Y, Zheng Y (2021) TransBridge: a lightweight transformer for left ventricle segmentation in echocardiography. In: Noble JA, Aylward S, Grimwood A, Min Z, Lee S-L, Hu Y (eds) Simplifying medical ultrasound, vol 12967. Springer, Cham, pp 63–72. https://doi.org/10.1007/978-3-030-87583-1_7
Jafari MH, Girgis H, Van Woudenberg N, Moulson N, Luong C, Fung A, Balthazaar S, Jue J, Tsang M, Nair P, Gin K, Rohling R, Abolmaesumi P, Tsang T (2020) Cardiac point-of-care to cart-based ultrasound translation using constrained CycleGAN. Int J Comput Assist Radiol Surg 15(5):877–886. https://doi.org/10.1007/s11548-020-02141-y
Escobar M, Castillo A, Romero A, Arbeláez P (2020) UltraGAN: ultrasound enhancement through adversarial generation. In: Burgos N, Svoboda D, Wolterink JM, Zhao C (eds) Simulation and synthesis in medical imaging, vol 12417. Springer, Cham, pp 120–130. https://doi.org/10.1007/978-3-030-59520-3_13
Saeed M, Muhtaseb R, Yaqub M (2022) Is contrastive learning suitable for left ventricular segmentation in echocardiographic images? https://doi.org/10.48550/ARXIV.2201.07219. Publisher: arXiv Version Number: 1
Zhu J-Y, Park T, Isola P, Efros AA (2020) Unpaired image-to-image translation using cycle-consistent adversarial networks. arXiv:1703.10593 [cs]
Isola P, Zhu J-Y, Zhou T, Efros AA (2018) Image-to-image translation with conditional adversarial networks. arXiv:1611.07004 [cs]. arXiv: 1611.07004
Chen L-C, Papandreou G, Schroff F, Adam H (2017) Rethinking atrous convolution for semantic image segmentation. arXiv:1706.05587 [cs]. arXiv: 1706.05587
Chen T, Kornblith S, Norouzi M, Hinton G (2020) A simple framework for contrastive learning of visual representations. In: International conference on machine learning. PMLR, pp 1597–1607
Grill J-B, Strub F, Altché F, Tallec C, Richemond P, Buchatskaya E, Doersch C, Avila Pires B, Guo Z, Gheshlaghi Azar M et al (2020) Bootstrap your own latent-a new approach to self-supervised learning. Adv Neural Inf Process Syst 33:21271–21284
Li M, Wang C, Zhang H, Yang G (2020) MV-RAN: multiview recurrent aggregation network for echocardiographic sequences segmentation and full cardiac cycle analysis. Comput Biol Med 120:103728. https://doi.org/10.1016/j.compbiomed.2020.103728
Wu H, Liu J, Xiao F, Wen Z, Cheng L, Qin J (2022) Semi-supervised segmentation of echocardiography videos via noise-resilient spatiotemporal semantic calibration and fusion. Med Image Anal 78:102397. https://doi.org/10.1016/j.media.2022.102397
Moal O, Roger E, Lamouroux A, Younes C, Bonnet G, Moal B, Lafitte S (2022) Explicit and automatic ejection fraction assessment on 2D cardiac ultrasound with a deep learning-based approach. Comput Biol Med 146:105637. https://doi.org/10.1016/j.compbiomed.2022.105637
Painchaud N, Duchateau N, Bernard O, Jodoin P-M (2022) Echocardiography segmentation with enforced temporal consistency. IEEE Trans Med Imaging. https://doi.org/10.1109/TMI.2022.3173669
Sirjani N, Moradi S, Oghli MG, Hosseinsabet A, Alizadehasl A, Yadollahi M, Shiri I, Shabanzadeh A (2022) Automatic cardiac evaluations using a deep video object segmentation network. Insights Imaging 13(1):69. https://doi.org/10.1186/s13244-022-01212-9
Ono S, Komatsu M, Sakai A, Arima H, Ochida M, Aoyama R, Yasutomi S, Asada K, Kaneko S, Sasano T, Hamamoto R (2022) Automated endocardial border detection and left ventricular functional assessment in echocardiography using deep learning. Biomedicines 10(5):1082. https://doi.org/10.3390/biomedicines10051082
Li M, Dong S, Gao Z, Feng C, Xiong H, Zheng W, Ghista D, Zhang H, de Albuquerque VHC (2020) Unified model for interpreting multi-view echocardiographic sequences without temporal information. Appl Soft Comput 88:106049. https://doi.org/10.1016/j.asoc.2019.106049
Shen Y, Zhang H, Fan Y, Lee AP, Xu L (2021) Smart health of ultrasound telemedicine based on deeply represented semantic segmentation. IEEE Internet Things J 8(23):16770–16778. https://doi.org/10.1109/JIOT.2020.3029957
Zyuzin V, Mukhtarov A, Neustroev D, Chumarnaya T (2020) Segmentation of 2D echocardiography images using residual blocks in U-net architectures. In: 2020 ural symposium on biomedical engineering, radioelectronics and information technology (USBEREIT), pp 499–502. IEEE, Yekaterinburg, Russia. https://doi.org/10.1109/USBEREIT48449.2020.9117678
Russakovsky O, Deng J, Su H, Krause J, Satheesh S, Ma S, Huang Z, Karpathy A, Khosla A, Bernstein M, Berg AC, Fei-Fei L (2015) ImageNet large scale visual recognition challenge. Int J Comput Vis 115(3):211–252. https://doi.org/10.1007/s11263-015-0816-y
Yakubovskiy P (2019) Segmentation models. GitHub
Sha Y (2021) Keras-unet-collection. GitHub. https://doi.org/10.5281/zenodo.5449801
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv preprint arXiv:1412.6980
Wang J, Zhu H, Wang S-H, Zhang Y-D (2021) A review of deep learning on medical image analysis. Mob Netw Appl 26(1):351–380. https://doi.org/10.1007/s11036-020-01672-7
Qin Z, Yu F, Liu C, Chen X (2018) How convolutional neural network see the world-a survey of convolutional neural network visualization methods. arXiv preprint arXiv:1804.11191
Chaurasia A, Culurciello E (2017) LinkNet: exploiting encoder representations for efficient semantic segmentation. 2017 IEEE visual communications and image processing (VCIP), pp 1–4. https://doi.org/10.1109/VCIP.2017.8305148. arXiv: 1707.03718
He K, Zhang X, Ren S, Sun J (2016) Deep residual learning for image recognition. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 770–778
Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B, Glocker B, Rueckert D (2018) Attention U-Net: learning where to look for the pancreas. arXiv:1804.03999 [cs]. arXiv: 1804.03999
Abraham N, Khan NM (2019) A novel focal tversky loss function with improved attention u-net for lesion segmentation. In: 2019 IEEE 16th international symposium on biomedical imaging (ISBI 2019). IEEE, pp 683–687
Chen J, Lu Y, Yu Q, Luo X, Adeli E, Wang Y, Lu L, Yuille AL, Zhou Y (2021) TransUNet: transformers make strong encoders for medical image segmentation. arXiv:2102.04306 [cs]. arXiv: 2102.04306
Simonyan K, Zisserman A (2014) Very deep convolutional networks for large-scale image recognition. arXiv preprint arXiv:1409.1556
Huang G, Liu Z, Van Der Maaten L, Weinberger KQ (2017) Densely connected convolutional networks. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 4700–4708
Dice LR (1945) Measures of the amount of ecologic association between species. Ecology 26(3):297–302
Real R, Vargas JM (1996) The probabilistic basis of Jaccard’s index of similarity. Syst Biol 45(3):380–385
Folland ED, Parisi AF, Moynihan PF, Jones DR, Feldman CL, Tow DE (1979) Assessment of left ventricular ejection fraction and volumes by real-time, two-dimensional echocardiography. A comparison of cineangiographic and radionuclide techniques. Circulation 60(4):760–766. https://doi.org/10.1161/01.CIR.60.4.760
Bland JM, Altman DG (2010) Statistical methods for assessing agreement between two methods of clinical measurement. Int J Nurs Stud 47(8):931–936. https://doi.org/10.1016/j.ijnurstu.2009.10.001
Chen Z, Zeng Z, Shen H, Zheng X, Dai P, Ouyang P (2020) DN-GAN: denoising generative adversarial networks for speckle noise reduction in optical coherence tomography images. Biomed Signal Process Control 55:101632. https://doi.org/10.1016/j.bspc.2019.101632
Acknowledgements
The authors would like to thank the Directorate General of Scientific Research and Technological Development (Direction Générale de la Recherche Scientifique et du Développement Technologique, DGRSDT, Algeria www.dgrsdt.dz) for the financial assistance towards this research.
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation and analysis were performed by Hafida Belfilali and Frédéric Bousefsaf. The first draft of the manuscript was written by Hafida Belfilali and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Consent to participate
Not applicable.
Consent for publication
Not applicable
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Belfilali, H., Bousefsaf, F. & Messadi, M. Left ventricle analysis in echocardiographic images using transfer learning. Phys Eng Sci Med 45, 1123–1138 (2022). https://doi.org/10.1007/s13246-022-01179-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-022-01179-3