Abstract
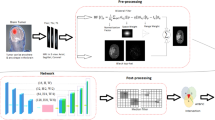
Automated assessment and segmentation of Brain MRI images facilitate towards detection of neurological diseases and disorders. In this paper, we propose an improved U-Net with VGG-16 to segment Brain MRI images and identify region-of-interest (tumor cells). We compare results of improved U-Net with a custom-designed U-Net architecture by analyzing the TCGA-LGG dataset (3929 images) from the TCI archive, and achieve pixel accuracies of 0.994 and 0.9975 from basic U-Net and improved U-Net architectures, respectively. Our results outperformed common CNN-based state-of-the-art works.





Similar content being viewed by others
References
Ronneberger O, Fischer P, Brox T (2015) U-net: convolutional networks for biomedical image segmentation. International conference on medical image computing and computer-assisted intervention. Springer, Cham, pp 234–241
Lin T-Y, Dollár P, Girshick R, He K, Hariharan B, Belongie S (2017) Feature pyramid networks for object detection. In: Proceedings of the IEEE conference on computer vision and pattern recognition, pp 2117–2125
Christ PF, Elshaer MEA, Ettlinger F, Tatavarty S, Bickel M, Bilic P, Rempfler M, Armbruster M, Hofmann F, D’Anastasi M, et al (2016) Automatic liver and lesion segmentation in CT using cascaded fully convolutional neural networks and 3D conditional random fields. In: International conference on medical image computing and computer-assisted intervention. Springer, pp 415–423
Menze BH, Jakab A, Bauer S, Kalpathy-Cramer Farahani K, Kirby J, Burren Y, Porz N, Slotboom J, Wiest R et al (2014) The multimodal brain tumor image segmentation benchmark (BRATS). IEEE Trans Med Imaging 34(10):1993–2024
Bengio Y, Courville A, Vincent P (2013) Representation learning: a review and new perspectives. IEEE Trans Pattern Anal Mach Intell 35(8):1798–1828
Havaei M, Davy A, Warde-Farley D, Biard A, Courville A, Bengio Y, Pal C, Jodoin P-M, Larochelle H (2017) Brain tumor segmentation with deep neural networks. Med Image Anal 35:18–31
Pereira S, Pinto A, Alves V, Silva CA (2016) Brain tumor segmentation using convolutional neural networks in MRI images. IEEE Trans Med Imaging 35(5):1240–1251
Kamnitsas K, Ledig C, Newcombe VFJ, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B (2017) Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal 36:61–78
Dvořák P, Menze B (2015) Local structure prediction with convolutional neural networks for multimodal brain tumor segmentation. In: International MICCAI workshop on medical computer vision. Springer, pp 59–71
Brosch T, Tang LYW, Yoo Y, Li DKB, Traboulsee A, Tam R (2016) Deep 3D convolutional encoder networks with shortcuts for multiscale feature integration applied to multiple sclerosis lesion segmentation. IEEE Trans Med Imaging 35(5):1229–1239
Dou Q, Chen HY, Lei LZ, Qin J, Wang D, Mok VCT, Shi L, Heng P-A (2016) Automatic detection of cerebral microbleeds from MR images via 3D convolutional neural networks. IEEE Trans Med Imaging 35(5):1182–1195
Maier O, Schröder C, Forkert ND, Martinetz T, Handels H (2015) Classifiers for ischemic stroke lesion segmentation: a comparison study. PLoS ONE 10(12):e0145118
Akkus Z, Ali I Sedlar J, Kline TL, Agrawal JP, Parney IF, Giannini C, Erickson BJ (2016) Predicting 1p19q chromosomal deletion of low-grade gliomas from mr images using deep learning. arXiv preprint. arXiv:1611.06939
Feng X, Tustison NJ, Patel SH, Meyer CH (2020) Brain tumor segmentation using an ensemble of 3D U-Nets and overall survival prediction using radiomic features. Front Comput Neurosci 14:25
Anaraki AK, Ayati M, Kazemi F (2019) Magnetic resonance imaging-based brain tumor grades classification and grading via convolutional neural networks and genetic algorithms. Biocybern Biomed Eng 39(1):63–74
Özyurt F, Sert E, Avci E, Dogantekin E (2019) Brain tumor detection based on convolutional neural network with neutrosophic expert maximum fuzzy sure entropy. Measurement 147:106830
Dolz J, Desrosiers C, Ayed IB (2018) IVD-Net: Intervertebral disc localization and segmentation in MRI with a multi-modal UNET. In: International workshop and challenge on computational methods and clinical applications for spine imaging, pp 130–143. Springer, pp 130–143
Lachinov D, Vasiliev E, Turlapov (2018) Glioma segmentation with cascaded UNET. In: International MICCAI Brainlesion workshop. Springer, pp 189–198
Hwang H, Ur Rehman HZ, Lee S (2019) 3D U-Net for skull stripping in brain MRI. Appl Sci 9(3):569
Kingma DP, Ba J (2014) Adam: a method for stochastic optimization. arXiv preprint. arXiv:1412.6980
Mazurowski MA, Clark K, Czarnek NM, Shamsesfandabadi P, Peters KB, Saha A (2017) Radiogenomics of lower-grade glioma: algorithmically-assessed tumor shape is associated with tumor genomic subtypes and patient outcomes in a multi-institutional study with the cancer genome atlas data. J Neuro-Oncol 133(1):27–35
Zhao L, Jia K (2015) Deep feature learning with discrimination mechanism for brain tumor segmentation and diagnosis. In: 2015 international conference on intelligent information hiding and multimedia signal processing (IIH-MSP). IEEE, pp 306–309
Funding
None.
Author information
Authors and Affiliations
Contributions
SG: Conceptualization, methodology and software; AC: Methodology and visualization; and KCS: Supervision, conceptualization, methodology, writing, reviewing and editing.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declared that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants performed by any of the authors.
Rights and permissions
About this article
Cite this article
Ghosh, S., Chaki, A. & Santosh, K. Improved U-Net architecture with VGG-16 for brain tumor segmentation. Phys Eng Sci Med 44, 703–712 (2021). https://doi.org/10.1007/s13246-021-01019-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13246-021-01019-w