Abstract
SIRT7, a sirtuin family member implicated in aging and disease, is a regulator of metabolism and stress responses. It remains elusive how human somatic stem cell populations might be impacted by SIRT7. Here, we found that SIRT7 expression declines during human mesenchymal stem cell (hMSC) aging and that SIRT7 deficiency accelerates senescence. Mechanistically, SIRT7 forms a complex with nuclear lamina proteins and heterochromatin proteins, thus maintaining the repressive state of heterochromatin at nuclear periphery. Accordingly, deficiency of SIRT7 results in loss of heterochromatin, de-repression of the LINE1 retrotransposon (LINE1), and activation of innate immune signaling via the cGAS-STING pathway. These aging-associated cellular defects were reversed by overexpression of heterochromatin proteins or treatment with a LINE1 targeted reverse-transcriptase inhibitor. Together, these findings highlight how SIRT7 safeguards chromatin architecture to control innate immune regulation and ensure geroprotection during stem cell aging.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sirtuins, mammalian homologs of the yeast longevity protein Sir2, are evolutionarily conserved nicotinamide adenine dinucleotide (NAD+)-dependent histone deacetylases (HDACs) involved in metabolic regulation and aging (Bishop and Guarente, 2007; Finkel et al., 2009; Herskovits and Guarente, 2013; Zhang et al., 2018). SIRT7 is the only sirtuin member that mainly localizes to the nucleolus (Michishita et al., 2005), a site for ribosome biogenesis and stress response (Shin et al., 2013; Tsai et al., 2014; Chen et al., 2016). Previous studies show that Sirt7-deficient mice have a shortened lifespan and suffer from aging-associated disorders (Vakhrusheva et al., 2008; Vazquez et al., 2016; Wronska et al., 2016). At the physiological level, SIRT7 has been implicated in hepatic lipid metabolism, cardiovascular system homeostasis and adipogenesis (Yoshizawa et al., 2014; Araki et al., 2015; Cioffi et al., 2015). At molecular levels, SIRT7 is broadly recognized for safeguarding genome integrity (Kiran et al., 2015; Vazquez et al., 2016; Paredes et al., 2018; Bao et al., 2019). For example, SIRT7 targets both acetylated H3 lysine 18 (Barber et al., 2012) and succinylated H3 lysine 122 (Li et al., 2016), and is recruited to DNA double-strand breaks (DSBs) in a PARP1-dependent manner, promoting chromatin condensation and DSB repair (Vazquez et al., 2016). Besides, SIRT7 is known as a direct transcriptional repressor that regulates mitochondrial and cytosolic protein homeostasis (Barber et al., 2012; Shin et al., 2013; Mohrin et al., 2015). Despite many seminal advances furthering our understanding of SIRT7 biology, the identification of reversible SIRT7-mediated cellular aging processes remains a challenge for the field.
Aging is a complicated process characterized by a progressive reduction in physiological integrity and organ function (Liu et al., 2012; Singh et al., 2019; He et al., 2020). A major contributor to organismal aging is stem cell senescence and exhaustion, which disrupts tissue maintenance and impairs organ regeneration (Ren et al., 2017). Human mesenchymal stem cells (hMSCs) support the maintenance of other cell populations in the body and have the potential to differentiate into diverse cell lineages, such as chondrocytes, osteoblasts, and adipocytes (Uccelli et al., 2008; Dimarino et al., 2013; Obeid et al., 2013). Not entirely surprisingly, progressive exhaustion of the hMSC pool has been causally linked to aging-associated tissue malfunction and degenerative diseases (Zhou et al., 2008; Zhang et al., 2011; Yang, 2018). In premature aging diseases like Hutchinson-Gilford progeria syndrome (HGPS), the hMSC population also undergoes accelerated decay with features of classic cellular senescence (Stenderup et al., 2003; Kudlow et al., 2007; Liu et al., 2011; Kubben et al., 2016; Wu et al., 2018). Small-molecule chemicals with geroprotective activity on hMSCs have exhibited efficacy to prolong healthspan and alleviate aging-associated syndromes in rodents (Geng et al., 2019). Thus, exploring molecular mechanisms of hMSC senescence can provide us with insight into how to ameliorate the development of age-related diseases or reverse cellular aging.
Cellular senescence is accompanied by marked epigenetic changes including altered DNA methylation patterns, aberrant histone modifications, and misleading nucleosome positioning (Talens et al., 2012; Lopez-Otin et al., 2013; Ren et al., 2017; Zhang et al., 2020). Heterochromatin disorganization, in particular, has been considered as a driver contributing to hMSC senescence in aging and diseases (Zhang et al., 2015; Deng et al., 2019). Heterochromatin endows particular genomic domains with enrichment of histone H3 Lys9 trimethylation (H3K9me3) and heterochromatin protein 1 α (HP1α) (Sridharan et al., 2013; Grewal and Jia, 2007; Kubben and Misteli, 2017). Such condensed heterochromatin prevents unequal recombination between repetitive regions and confers repression of transposable elements (TE) (Bourque et al., 2018; Zhang et al., 2020) that are primarily located at genomic lamina-associated domains (LADs) and spatially anchored to the nuclear periphery with nuclear lamina proteins (Guelen et al., 2008; van Steensel and Dekker, 2010; Bickmore and van Steensel, 2013). Emerging shreds of evidence have indicated that de-repression of retrotransposons such as long interspersed element 1 (LINE1) occurs during organismal and cellular aging and could be a new driver for cellular senescence (Garcia-Perez et al., 2010; Gorbunova et al., 2014; Van Meter et al., 2014; De Cecco et al., 2019; Simon et al., 2019). To date, the mechanisms underlining heterochromatin maintenance in human stem cells are largely unknown.
In this study, we discovered that SIRT7 functions to tether heterochromatin to the nuclear lamina and thereby maintains heterochromatin organization in hMSCs. We also found that SIRT7 downregulation associated with hMSC senescence accounts for detachment of LADs from the nuclear lamina, loss of heterochromatin, and de-repression of LINE1, events that activate the innate immune response through the cyclic GMP-AMP synthase-stimulator of interferon genes (cGAS-STING) signaling. Restoration of heterochromatin compaction or inhibition of LINE1 retrotransposition rescued the senescence phenotypes in SIRT7-deficient hMSCs. These results reveal that leveraging heterochromatin-innate immune response pathways is a novel facet by which SIRT7 safeguards hMSC integrity.
Results
SIRT7 is downregulated in senescent hMSCs
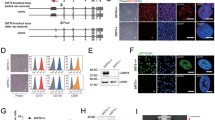
To evaluate SIRT7 expression in human cellular models of aging, we first measured the SIRT7 protein level in replicatively senescent (RS) hMSCs and HGPS-specific (LMNAG608G/+ or LMNAG608G/G608G) hMSCs, known to be senescent prematurely (Liu et al., 2011; Wu et al., 2018). Western blot analysis revealed that SIRT7 protein level was decreased in late-passage versus early-passage wild-type (WT, SIRT7+/+) hMSCs but also in HGPS hMSCs relative to control hMSCs (Fig. 1A and 1B). We also found that SIRT7 was downregulated in primary hMSCs derived from physiologically aged individuals relative to hMSCs derived from younger individuals (Figs. 1C and S1A). Collectively, these data suggest that the downregulation of SIRT7 is a phenotype of cellular aging in hMSCs.
Generation and characterization of SIRT7-deficient hESCs. (A) Left, Western blot analysis of SIRT7 protein in replicatively senescent (RS) hMSCs at early (EP, P3) and late passages (LP, P8) with β-Tubulin as loading control. Right, statistical analysis of relative SIRT7 protein expression levels. Data are presented as the means ± SEM. n = 3. *, P < 0.05 (t test). (B) Left, Western blot analysis of SIRT7 protein in WT and HGPS-specific (LMNAG608G/+ or LMNAG608G/G608G) hMSCs at LP (P8) with β-Tubulin used as loading control. Right, statistical analysis of relative SIRT7 protein expression levels. Data are presented as the means ± SEM. n = 3. *, P < 0.05, **, P < 0.01 (t test). (C) Statistical analysis of relative SIRT7 protein expression levels in young and old primary hMSCs. Data are presented as the means ± SEM. n = 4 samples. *, P < 0.05 (t test). (D) Left, schematic illustration of SIRT7 gene editing (exon 4) using CRISPR/Cas9-mediated non-homologous end joining (NHEJ) in hESCs. Right, DNA sequence chromatogram showing the introduction of termination codon TAA by gene editing. (E) Schematic workflow showing the generation of SIRT7+/+ and SIRT7−/− hMSCs from hESCs. (F) Genome-wide analysis of copy number variations (CNVs) in SIRT7+/+ and SIRT7−/− hMSCs at middle passage (MP, P6). (G) Western blot analysis of SIRT7 protein in SIRT7+/+ and SIRT7−/− hMSCs with β-Tubulin used as loading control. (H) Growth curves of SIRT7+/+ and SIRT7−/− hMSCs. Data are presented as the means ± SEM. n = 3. (I) SA-β-gal staining of SIRT7+/+ and SIRT7−/− hMSCs at EP (P3) and LP (P8). Scale bar, 125 μm. Data are presented as the means ± SEM. n = 3. ns, not significant, **, P < 0.01 (t test). (J) Clonal expansion analysis of SIRT7+/+ and SIRT7−/− hMSCs at EP (P3) and LP (P8). Data are presented as the means ± SEM. n = 3. ns, not significant, **, P < 0.01 (t test). (K) Immunostaining of Ki67 in SIRT7+/+ and SIRT7−/− hMSCs at EP (P3) and LP (P8). Scale bar, 25 μm. Data are presented as the means ± SEM. n = 3. **, P < 0.01 (t test). (L) Bar plot showing the percentages of cells in S-phase of cell cycle in SIRT7+/+ and SIRT7−/− hMSCs at EP (P3) and LP (P8). Data are presented as the means ± SEM. n = 3. **, P < 0.01, ***, P < 0.001 (t test). (M) ROS levels were determined by staining with the free radical sensor H2DCFDA and measured by FACS in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). Data are presented as the means ± SEM. n = 3. (N) Heatmap showing quantitative RT-PCR analysis of aging-related genes in SIRT7+/+ and SIRT7−/− hMSCs at EP (P3) and LP (P8) hMSCs. Expression levels of the indicated genes in each cell type were normalized to those in SIRT7+/+ hMSCs at EP (P3). (O) Western blot analysis of cyclin-dependent kinase inhibitors P16 and P21 proteins in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6) with GAPDH used as loading control. (P) Analysis of luciferase activities in TA muscles of immunodeficient mice transplanted with SIRT7+/+ (left) or SIRT7−/− hMSCs (right) at MP (P6) in Day 0, 3 and 5 after implantation. Data calculated by the ratios of log2 (SIRT7−/−/SIRT7+/+) are presented as the means ± SEM. n = 6. ns, not significant, ***, P < 0.001 (t test)
SIRT7 deficiency accelerates hMSC senescence
Using a CRISPR/Cas9-assisted gene knockout strategy with sgRNAs targeting SIRT7, we next generated SIRT7-deficient human embryonic stem cells (hESCs), which were homozygous for a mutation in SIRT7 resulting in premature termination of SIRT7 translation (Fig. 1D). Successful ablation of SIRT7 protein was verified with Western blot (Fig. S1B) while karyotyping and genome-wide copy number variation (CNV) analyses demonstrated that the genomic integrity of SIRT7-deficient (SIRT7−/−) hESCs remained intact (Fig. S1C and S1D). Also, we observed normal morphology and comparable expression of pluripotency markers NANOG, SOX2 and OCT4 in SIRT7−/− hESCs relative to SIRT7+/+ hESCs (Fig. S1E). No difference in proliferative potential was observed between SIRT7+/+ and SIRT7−/− hESCs (Fig. S1F). In all, these data suggest that SIRT7 is dispensable for maintaining hESC properties.
We then differentiated SIRT7+/+ and SIRT7−/− hESCs into hMSCs (referred to hereafter as SIRT7+/+ and SIRT7−/− hMSCs, respectively) (Fig. 1E). Both SIRT7+/+ and SIRT7−/− hMSCs were positive for typical hMSC surface markers including CD105, CD90, CD73, CD44, CD166 and CD29, and negative for non-hMSC markers, such as CD164, CD14, CD19 and podoplanin (PDPN) (Fig. S2A) (Secunda et al., 2015; Miura, 2016). Both types of hMSCs maintained chromosomal integrity as verified with CNV analysis (Fig. 1F), and the absence of SIRT7 protein (Fig. 1G) had no noticeable effect on hMSC differentiation into osteoblasts, chondrocytes, and adipocytes (Fig. S2B).
To investigate whether SIRT7 deficiency accelerates hMSC senescence, we subjected SIRT7+/+ and SIRT7−/− hMSCs to serial passaging. Compared with SIRT7+/+ hMSCs, which exhibited normal aging kinetics, SIRT7−/− hMSCs underwent complete growth arrest as early as by passage 10 (Fig. 1H). We also observed increased percentages of senescence-associated β-galactosidase (SA-β-gal)-positive cells (Fig. 1I), compromised clonal expansion ability (Fig. 1J), decreased ratios of Ki67-positive cells (Fig. 1K) and reduced percentages of S-phase cells in SIRT7−/− relative to SIRT7+/+ hMSCs (Fig. 1L). In addition, SIRT7 deficiency resulted in higher reactive oxygen species (ROS) levels and an increased DNA damage response (DDR) (Figs. 1M, S2C and S2D). Consistent with the accelerated senescence phenotypes, we observed the upregulation of cyclin-dependent kinase inhibitors and aging markers CDKN1A (P21) and CDKN2A (P16) at both mRNA and protein levels, along with transcriptional downregulation of LMNB1 and TMPO in SIRT7−/− hMSCs (Fig. 1N and 1O). RNA sequencing (RNA-seq) results further showed that genes differentially expressed between SIRT7−/− and SIRT7+/+ hMSCs were primarily associated with the terms “aging” and “senescence-associated secretory phenotype (SASP)” (Fig. S2E–G). Finally, SIRT7−/− hMSCs underwent accelerated decay in vivo when implanted into the tibialis anterior (TA) muscles of immunodeficient mice relative to SIRT7+/+ hMSCs (Fig. 1P).
To confirm a direct role for SIRT7 in hMSC senescence, we transduced WT hMSCs with lentiviruses encoding CRISPR/Cas9 and SIRT7-targeted sgRNAs (Sanjana et al., 2014) (Fig. S2H). Similar to what we had observed in SIRT7-null hESC-derived hMSCs, SIRT7 ablation resulted in increased percentages of SA-β-gal-positive cells and decreased clonal expansion ability, reflecting accelerated senescence (Fig. S2I and S2J). Thus, these data suggest that SIRT7 deficiency accelerates functional attrition of hMSCs.
SIRT7 stabilizes heterochromatin in hMSCs
SIRT7 was initially identified as a histone deacetylase capable of removing acetyl groups from acetylated H3K18 and H3K36 in cancer cells and more recently reported as a histone desuccinylase for H3K122 residues across commonly used cell lines (Barber et al., 2012; Li et al., 2016; Wang et al., 2019a). We examined these typical histone modification substrates in SIRT7−/− hMSCs but did not find that SIRT7 depletion resulted in any detectable changes of these substrates (Fig. S2K).
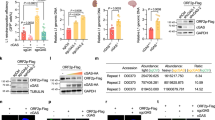
To investigate a histone deacetylase or desuccinylase activity-independent role for SIRT7 in the regulation of hMSC homeostasis, we sought to identify additional interaction partners of SIRT7. To this end, we expressed Flag-tagged SIRT7 proteins in HEK293T cells and performed immunoprecipitation (IP) with an anti-Flag antibody followed by liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis (Fig. 2A and 2B). In addition to previously identified SIRT7-interacting proteins such as fibrillarin (FBL) (Iyer-Bierhoff et al., 2018), we identified a list of candidate SIRT7-interacting proteins associated with heterochromatin organization, such as nuclear lamina protein LBR and heterochromatin protein KAP1 (Fig. S3A). Furthermore, by performing co-immunoprecipitation (co-IP) assay, we identified additional nuclear lamina proteins Lamin B1 and Emerin, as well as heterochromatin proteins HP1α and HP1γ as novel SIRT7-interacting proteins (Fig. 2C and 2D). In addition, Western blot and immunofluorescence analyses revealed decreased levels of these nuclear lamina proteins (i.e., LAP2β) and heterochromatin marks (i.e., HP1α and H3K9me3) in SIRT7−/− hMSCs relative to levels in SIRT7+/+ hMSCs (Figs. 2E–H and S3B). Consistently, we observed an increased degree of heterochromatin loss at the nuclear periphery in nuclei of SIRT7−/− hMSCs under an electron microscope (Fig. S3C). Together, these data indicate that SIRT7 is required for maintaining heterochromatin in hMSCs.
SIRT7 interacts with nuclear lamina proteins and heterochromatin proteins. (A) Schematic of mass spectrometry work flow for identifying SIRT7-interacting proteins. Luc was used as control. (B) Gene Ontology Cellular Component (GO-CC) enrichment analysis of candidate SIRT7-interacting proteins identified by mass spectrometry. (C) Co-immunoprecipitation analysis of LBR, Lamin B1, Emerin, KAP1, HP1α and HP1γ with exogenous Flag-tagged SIRT7 protein in HEK293T cells. (D) Co-immunoprecipitation analysis of LBR, Lamin B1, Emerin, KAP1, HP1α and HP1γ with endogenous SIRT7 protein in WT hMSCs. The band corresponding to LBR is indicated with an asterisk. (E) Left, Western blot analysis of heterochromatin-related proteins in hMSCs at MP (P6) with β-Tubulin used as loading control. Right, statistical analysis of the relative heterochromatin-related protein expression levels. Data are presented as the means ± SEM. n = 3. *, P < 0.05 (t test). (F) Left, immunostaining of HP1α and Lamin A/C in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). White arrowheads indicate cells with decreased expression of HP1α. Right, mean fluorescence intensity of HP1α was measured by Image J. Scale bar, 25 μm. Data are presented as the means ± SEM. n = 100 cells. ***, P < 0.001 (t test). (G) Left, immunostaining of LAP2β in SIRT7+/+ and SIRT7−/− hMSCs. White arrowheads indicate cells with decreased expression of LAP2β. Right, mean fluorescence intensity of LAP2β was measured by Image J. Scale bar, 25 μm. Data are presented as the means ± SEM. n = 150 cells. ***, P < 0.001 (t test). (H) Left, z-stack 3D reconstruction of H3K9me3 and Lamin A/C immunofluorescence images (shown in Fig. S3B) in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). Scale bar, 5 μm. Right, mean fluorescence intensity of H3K9me3 in Fig. S3B was quantified with Image J. Data are presented as means ± SEM. n = 150 cells. ***, P < 0. 001 (t test)
To characterize the heterochromatin state managed by SIRT7 in greater detail, we performed DNA adenine methyltransferase identification with high-throughput sequencing (DamID-seq) that is a powerful tool to study the interactions between nuclear lamina and chromatin (Guelen et al., 2008), H3K9me3 chromatin immunoprecipitation followed by high-throughput sequencing (ChIP-seq), and chromatin accessibility assay (Assay for transposase accessible chromatin sequencing, ATAC-seq) in SIRT7+/+ and SIRT7−/− hMSCs. DamID was performed by introducing DNA adenine methyltransferase (Dam) fused to Emerin (a nuclear lamina-associated protein), or Dam protein alone into SIRT7+/+ and SIRT7−/− hMSCs to map the changes in LADs upon SIRT7 ablation (Fig. 3A). SIRT7 deficiency resulted in a decrease of DamID signals (Figs. 3B, 3C, and S4A–C), and the DamID signals in LAD-localized repetitive elements (LINE, SINE, LTR, Satellite and rRNA) were all diminished in SIRT7−/− hMSCs (Fig. 3D). As expected, ChIP-seq analysis demonstrated reduced enrichment in long-range H3K9me3 occupancy (H3K9me3 mountains) in constitutive heterochromatin regions, especially at the LAD regions and the LAD located repetitive elements, indicative of a reduced association between heterochromatin and nuclear lamina (Figs. 3E–G and S4D–I). Consistent with the global heterochromatin decondensation observed in SIRT7−/− hMSCs, ATAC-seq revealed that heterochromatin regions became more accessible when SIRT7 was depleted (Figs. 3H, 3I, and S4J–M). These data indicate a key role of SIRT7 in maintaining heterochromatin at the nuclear periphery.
SIRT7 is required for heterochromatin maintenance in hMSCs. (A) Schematic diagram showing the strategy for DamID-seq library construction. When the Dam-Emerin fused protein was expressed in hMSCs, the genomic DNA region near the nuclear envelope could be methylated by DNA adenine methyltransferase (Dam) at adenines. A parallel experiment “Dam only” was used to eliminate the background Dam signal. After genome extraction, the sequence containing methylated sites could be specifically cut by DpnI and amplified by PCR. The amplified fragments were then proceeded to DNA library construction and high-throughput sequencing. (B) Chromosome ideogram showing relative DamID signals in LADs across 23 chromosomes at MP (P6). The color key from blue to red shows low to high relative DamID levels, respectively. (C) Violin plot showing the DamID signal [log2 (Dam-EMD/Dam)] in LADs in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). The white circles represent the median values, and the white lines represent the values within the IQR from smallest to largest. ***, P < 0.001 (Two-sided Wilcoxon rank-sum test). (D) Violin plot showing the DamID signal [log2 (Dam-EMD/ Dam)] in LADs located in repetitive elements, including SINE, LINE, LTR, Satellite, rRNA, low complexity and simple repeat elements, in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). The white circles represent the median values, and the white lines represent the values within the IQR from smallest to largest. ***, P < 0.001 (Two-sided Wilcoxon rank-sum test). (E) Chromosome ideogram showing the relative H3K9me3 signal in “H3K9me3 mountains” across 23 chromosomes at MP (P6). The color key from blue to red shows low to high relative H3K9me3 levels, respectively. (F) Violin plot showing the H3K9me3 signal in “H3K9me3 mountains” in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). The white circles represent median values, and the white lines represent the values within the IQR from smallest to largest. ***, P < 0.001 (Two-sided Wilcoxon rank-sum test). (G) Violin plot showing the H3K9me3 signal in “H3K9me3 mountains” located in repetitive elements, including SINE, LINE, LTR, Satellite, rRNA, low complexity and simple repeat elements in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). The white circles represent the median values, and the white lines represent the values within the IQR from smallest to largest. **, P < 0.01, ***, P < 0.001 (Two-sided Wilcoxon rank-sum test). (H) Metaplots showing the average ATAC signals for all ATAC peaks, ATAC peaks in LADs, and ATAC peaks in “H3K9me3 mountains” in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). ***, P < 0.001 (Two-sided Wilcoxon rank-sum test). (I) Heatmap showing the relative enrichment of ATAC peaks in repetitive elements, including SINE, LINE, LTR, Satellite, rRNA, low complexity and simple repeat elements in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). Enrichment of ATAC peaks in repetitive elements in SIRT7−/− hMSCs were normalized to those in SIRT7+/+ hMSCs
SIRT7 deficiency activates LINE1 in hMSCs
Constitutive heterochromatin is normally required for the inactivation of retrotransposons such as LINE1 (De Cecco et al., 2013; Gorbunova et al., 2014; Van Meter et al., 2014). Using ChIP-qPCR assay, we detected that SIRT7 bound specific LINE1 promoter regions in SIRT7+/+ but not in SIRT7−/− hMSCs (Fig. 4A). As we observed an increase in accessibility at a variety of genomic repetitive elements, ChIP-qPCR indicated a decrease in H3K9me3 enrichment at LINE1 promoter when SIRT7 was absent (Fig. 4B). The combined results from DamID-seq, H3K9me3 ChIP-seq, and ATAC-seq analyses supported the notion that deficiency of SIRT7 in hMSCs led to de-repression and increased chromatin accessibility of LINE1 (Fig. 4C–E). Real-time quantitative PCR (RT-qPCR) indeed demonstrated increased RNA levels of LINE1 in SIRT7−/− hMSCs (Fig. 4F). Consistent with the elevated transcription activity of LINE1 in SIRT7−/− hMSCs, we found that LINE1 coding proteins ORF1 and ORF2 were upregulated both in SIRT7−/− hMSCs and physiologically senescent primary hMSCs (Figs. 4G and S5A). To examine whether the retrotransposition activity of LINE1 is increased in the absence of SIRT7, we conducted retrotransposition analysis in SIRT7+/+ and SIRT7−/− hMSCs with an engineered LINE1 reporter system (Thomas et al., 2017). As shown in Fig. 4H, a higher retrotransposition frequency of LINE1 was observed in SIRT7−/− hMSCs relative to SIRT7+/+ hMSCs. We also detected an elevated genomic DNA content of LINE1 in SIRT7−/− hMSCs with RT-qPCR (Fig. 4I). These results indicate that SIRT7 deficiency enables increased transcription of LINE1 and subsequent LINE1-dependent retrotransposition events.
Depletion of SIRT7 in hMSCs induces LINE1 activation and triggers innate immune responses via cGAS-STING pathway. (A) ChIP-qPCR assessment of SIRT7 enrichment at seven LINE1 regions in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). Data are presented as the means ± SEM. n = 3. **, P < 0.01. ***, P < 0.001 (t test). (B) ChIP-qPCR assessment of H3K9me3 enrichment of LINE1 regions in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6) using the fourth LINE1 primer. Data are presented as the means ± SEM. n = 4. ***, P < 0.001 (t test). (C) Violin plot showing the DamID signal [log2 (Dam-EMD/ Dam)] in LINE1 regions located in LADs in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). The white circles represent the median values, and the white lines represent the values within the IQR from smallest to largest. ***, P < 0.001 (Two-sided Wilcoxon rank-sum test). (D) Violin plot showing the H3K9me3 signal in LINE1 regions located in “H3K9me3 mountains” in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). The white circles represent the median values, and the white lines represent the values within the IQR from smallest to largest. ***, P < 0.001 (Two-sided Wilcoxon rank-sum test). (E) Metaplot showing the average ATAC signal for ATAC peaks in LINE1 regions in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). ***, P < 0.001 (Two-sided Wilcoxon rank-sum test). (F) Quantitative RT-PCR analysis of LINE1 transcript levels in SIRT7+/+ and SIRT7−/− hMSCs at EP (P3) and LP (P8). Data are presented as the means ± SEM. n = 4. *, P < 0.05. ***, P < 0.001 (t test). (G) Left, Western blot analysis of LINE1 ORF1 and ORF2 proteins in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6) with H3 used as loading control. Right, statistical analysis of relative ORF1 and ORF2 protein expression levels. Data are presented as the means ± SEM. n = 3. *, P < 0.05, **, P < 0.01 (t test). (H) Left, schematic of retrotransposition assay. The plasmid p99-GFP-LRE3-Cherry lacking a CMV promoter encodes a full-length LINE1 element tagged with an indicator cassette of mEGFP1 retrotransposition and mCherry cassette. Right, quantification of de novo retrotransposition events (EGFP-positive cells) in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6), normalized by live cell numbers and transfection efficiency. Data are presented as the means ± SEM. n = 3. *, P < 0.05. ***, P < 0.001 (t test). (I) RT-qPCR analysis of the relative LINE1 genomic DNA content in SIRT7+/+ and SIRT7−/− hMSCs at EP (P3) and LP (P8). Data are presented as the means ± SEM. n = 4. *, P < 0.05. ***, P < 0.001 (t test). (J) Western blot analysis of the phosphorylation levels of P65, TBK1 and IRF3 in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6) with GAPDH used as loading control. (K) ELISA analysis of IL6 and MCP1 levels in culture medium of SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). IL6 levels were normalized to cell number. Data are presented as the means ± SEM. n = 5. MCP1 levels were normalized to cell number. n = 3. **, P < 0.001. ***, P < 0.001 (t test). (L) Heatmap showing quantitative RT-PCR analysis of IFN-I and SASP genes in SIRT7+/+ and SIRT7−/− hMSCs at MP (P6). Expression levels of these indicated genes in SIRT7−/− hMSCs were normalized to those in SIRT7+/+ hMSCs
Activation of cGAS-STING pathway and innate immune responses in SIRT7-deficient hMSCs
Given reports on LINE1 activation functioning as a fuse for the DNA sensing cGAS-STING signaling pathway (Volkman and Stetson, 2014; De Cecco et al., 2019; Geng et al., 2019; Simon et al., 2019), we next interrogated its potential association with SIRT7 deficiency and activation of LINE1. Activation of the cGAS-STING pathway was evident in SIRT7-deficient hMSCs, as detected by increased levels of phosphorylated forms of NF-κB P65, TANK-binding kinase 1 (TBK1) and IFN regulatory factor 3 (IRF3) (Fig. 4J). Correspondingly, elevated gene expression related to type-I interferon (IFN-I) and SASP responses, as downstream effectors of cGAS-STING signaling was observed in SIRT7−/− hMSCs (Figs. 4K, 4L, and S2G), indicating that SIRT7 deficiency contributes to the appearance of a range of senescence-related phenotypes, at least in part by activation of LINE1 events and the cGAS-STING pathway.
SIRT7 can be targeted to rejuvenate aged hMSCs
To investigate how the epistasis between SIRT7, heterochromatin loss, LINE1 activation, and induction of the cGAS-STING pathway coordinate acquisition of hMSC senescence, we performed the following experiments. First, we restored the expression of SIRT7 in SIRT7−/− hMSCs via lentiviral vector-mediated gene transfer. As expected, re-introduction of SIRT7 restored H3K9me3 levels, reduced LINE1 transcription, and attenuated IFN-I and SASP responses, and rescued phenotypes of accelerated hMSC senescence. We observed decreased percentages of SA-β-gal-positive cells, increased clonal expansion ability, and a increase in Ki67-positive proliferating cells (Figs. 5A–D, S5B, and S5C). Second, considering that nuclear lamina proteins (i.e., Lamin B1) and heterochromatin proteins (i.e., KAP1 and HP1α) as newly identified SIRT7 interaction partners were downregulated in SIRT7−/− hMSCs, we examined how ectopic expression of Lamin B1 and heterochromatin proteins KAP1 and HP1α affected SIRT7-deficient cells. Similar to re-introduction of SIRT7, restoring Lamin B1, KAP1 or HP1α expression in SIRT7−/− hMSCs alleviated aging defects, including decreased percentages of SA-β-gal-positive cells, increased proliferative potential, and attenuated expression of IFN-I and SASP genes (Figs. 5E–G, S5D, and S5E). Third, we asked whether blocking the cGAS-STING pathway would rescue the senescence phenotypes caused by SIRT7 deficiency. shRNA knockdown of STING in SIRT7−/− hMSCs led to alleviation of the senescence phenotypes, resembling overexpression of SIRT7 or individual heterochromatin components (Figs. 5H–K and S5F). Collectively, these data converge on a SIRT7-heterochromatin-cGAS-STING axis as an important geroprotective mechanism in hMSCs.
SIRT7-heterochromatin-cGAS-STING axis protects hMSCs against senescence. (A) Left, immunostaining of H3K9me3 and Lamin A/C in SIRT7−/− hMSCs transduced with lentiviruses expressing Luc or SIRT7. White arrowheads indicate cells with decreased expression of H3K9me3. Scale bar, 25 μm. Right, mean fluorescence intensity of H3K9m3 was measured by Image J. Data are presented as the means ± SEM. n = 150 cells. ***, P < 0.001 (t test). (B) Quantitative RT-PCR analysis of LINE1 transcript levels in SIRT7−/− hMSCs transduced with lentiviruses expressing Luc or SIRT7. Data are presented as the means ± SEM. n = 4. ***, P < 0.001 (t test). (C) Heatmap showing quantitative RT-PCR analysis of IFN-I and SASP genes in SIRT7−/− hMSCs transduced with lentiviruses expressing Luc or SIRT7. Expression levels of these indicated genes in SIRT7 lentivirus-transduced hMSCs were normalized to those in Luc lentivirus-transduced hMSCs. (D) SA-β-gal staining of SIRT7−/− hMSCs transduced with lentiviruses expressing Luc or SIRT7. Scale bar, 125 μm. Data are presented as means ± SEM. n = 3. *, P < 0.05 (t test). (E) SA-β-gal staining of SIRT7−/− hMSCs transduced with lentiviruses expressing KAP1, HP1α or Lamin B1. Scale bar, 125 μm. Data are presented as the means ± SEM. n = 3. **, P < 0.01 (t test). (F) Clonal expansion analysis of SIRT7−/− hMSCs transduced with lentiviruses expressing KAP1, HP1α or Lamin B1. Data are presented as the means ± SEM. n = 3. *, P < 0.05 (t test). (G) Heatmap showing quantitative RT-PCR analysis of IFN-I and SASP genes in physiologically senescent hMSCs from old donor transduced with lentiviruses expressing KAP1, HP1α or Lamin B1. Expression levels of these indicated genes in KAP1, HP1α or Lamin B1-transduced hMSCs were normalized to those in Luc-transduced hMSCs. (H) SA-β-gal staining of SIRT7−/− hMSCs transduced with lentiviruses expressing sh-GL2 or sh-STING. Scale bar, 125 μm. Data are presented as the means ± SEM. n = 3. **, P < 0.01 (t test). (I) Clonal expansion analysis of SIRT7−/− hMSCs transduced with lentiviruses expressing sh-GL2 or sh-STING. Data are presented as the means ± SEM. n = 3. **, P < 0.01 (t test). (J) Immunostaining of Ki67 in SIRT7−/− hMSCs transduced with lentiviruses expressing sh-GL2 or sh-STING. White arrowheads indicate cells with decreased expression of Ki67. Scale bar, 25 μm. Data are presented as the means ± SEM. n = 3. ***, P < 0.001 (t test). (K) Heatmap showing quantitative RT-PCR analysis of IFN-I and SASP genes in SIRT7−/− hMSCs transduced with lentiviruses expressing sh-GL2 or sh-STING. Expression levels of these indicated genes in sh-STING lentivirus-transduced hMSCs were normalized to those in sh-GL2 lentivirus-transduced hMSCs. (L) SA-β-gal staining of RS hMSCs transduced with lentiviruses expressing Luc or SIRT7. Scale bar, 125 μm. Data are presented as the means ± SEM. n = 3. ***, P < 0.001 (t test). (M) Clonal expansion analysis of RS hMSCs transduced with lentiviruses expressing Luc or SIRT7. Data are presented as the means ± SEM. n = 3. *, P < 0.05 (t test). (N) Immunostaining of Ki67 in RS hMSCs transduced with lentiviruses expressing Luc or SIRT7. White arrowheads indicate cells with decreased expression of Ki67. Scale bar, 25 μm. Data are presented as the means ± SEM. n = 3. *, P < 0.05 (t test). (O) Heatmap showing quantitative RT-PCR analysis of IFN-I and SASP genes in RS hMSCs transduced with lentiviruses expressing Luc or SIRT7. Expression levels of these indicated genes in SIRT7 lentivirus-transduced hMSCs were normalized to those in Luc lentivirus-transduced hMSCs. (P) SA-β-gal staining of old primary hMSCs from an 80-year-old individual (Old #2) transduced with lentiviruses expressing Luc or SIRT7. Scale bar, 125 μm. Data are presented as the means ± SEM. n = 3. **, P < 0.01 (t test). (Q) Clonal expansion analysis of old primary hMSCs from an 80-year-old individual (Old #2) transduced with lentiviruses expressing Luc or SIRT7. Data are presented as the means ± SEM. n = 5. ***, P < 0.001 (t test). (R) Immunostaining of Ki67 in old primary hMSCs from an 80-year-old individual (Old #2) transduced with lentiviruses expressing Luc or SIRT7. White arrowheads indicate cells with decreased expression of Ki67. Scale bar, 25 μm. Data are presented as the means ± SEM. n = 3. ***, P < 0.001 (t test). (S) Heatmap showing quantitative RT-PCR analysis of IFN-I and SASP genes in old primary hMSCs from an 80-year-old individual (Old #2) transduced with lentiviruses expressing Luc or SIRT7. Expression levels of these indicated genes in SIRT7 lentivirus-transduced hMSCs were normalized to those in Luc lentivirus-transduced hMSCs
Finally, we explored whether targeting SIRT7 might rejuvenate replicatively and physiologically senescent primary hMSCs isolated from an 80-year-old individual. Indeed, we found that restoring SIRT7 similarly alleviated the senescence phenotypes we had observed in SIRT7-null hMSCs, including decreased percentages of SA-β-gal-positive cells, improved cellular expansion ability, increased Ki67-positive cells, and attenuated expression of IFN-I and SASP genes (Fig. 5L–S).
3TC is a geroprotective compound for hMSCs
Reverse-transcriptase inhibitors (RTis) are nucleoside analogs that can inhibit reverse transcriptase, among which the anti-HIV1 drug Lamivudine (3TC) has been shown to inhibit reverse transcription of LINE1 (Jones et al., 2008; Dai et al., 2011). We, therefore, wondered whether 3TC-mediated reduction of the LINE1 life cycle might rescue senescence phenotypes in SIRT7−/− hMSCs. The treatment of SIRT7−/− hMSCs with 10 μmol/L 3TC led to the downregulation of the LINE1 genomic DNA content (Fig. 6A), and rescued cellular senescence phenotypes, as detected by reduced percentages of SA-β-gal-positive cells, increased proliferative potential, increased percentages of S-phase cells, upregulated transcript levels of LMNB1 and TMPO, reduced expression of aging markers CDKN1A (P21) and CDKN2A (P16), and reduced aging-associated immune activation (as detected by reduced IFN-I and SASP responses) (Fig. 6B–G). More importantly, 3TC treatment alleviated senescence phenotypes in physiologically senescent hMSCs isolated from aged individuals (Fig. 6H). Our findings reveal a critical role of the cGAS-STING pathway in mediating geroprotection downstream of SIRT7, but also point to the possibility of developing 3TC as a therapy for alleviating stem cell senescence and treating aging-related disorders (Fig. 6I).
3TC attenuates SIRT7-deficient hMSCs senescence by antagonizing LINE1’s effects. (A) RT-qPCR analysis of the relative LINE1 genomic DNA content in SIRT7−/− hMSCs treated with vehicle or 3TC. Data are presented as the means ± SEM. n = 4. *, P < 0.05 (t test). (B) SA-β-gal staining of SIRT7−/− hMSCs treated with vehicle and 3TC. Scale bar, 125 μm. Data are presented as means ± SEM. n = 3. ***, P < 0.001 (t test). (C) Clonal expansion analysis of SIRT7−/− hMSCs treated with vehicle and 3TC. Data are presented as means ± SEM. n = 3. **, P < 0.01 (t test). (D) Bar plot showing the percentages of cells in S-phase of cell cycle in SIRT7−/− hMSCs treated with vehicle or 3TC. Data are presented as the means ± SEM. n = 3. ***, P < 0.001 (t test). (E) Heatmap showing quantitative RT-PCR analysis of aging-related markers in SIRT7−/− hMSCs treated with vehicle or 3TC. Expression levels of these indicated genes in 3TC-treated hMSCs were normalized to those in vehicle-treated hMSCs. (F) Western blot analysis of P16 and P21 proteins in SIRT7−/− hMSCs treated with vehicle or 3TC with β-Tubulin used as loading control. (G) Heatmap showing quantitative RT-PCR analysis of IFN-I and SASP genes in SIRT7−/− hMSCs treated with vehicle or 3TC. Expression levels of these indicated genes in 3TC-treated hMSCs were normalized to those in vehicle-treated hMSCs. (H) SA-β-gal staining of old primary hMSCs from an 80-year-old individual (Old #2) treated with vehicle or 3TC. Scale bar, 125 μm. Data are presented as means ± SEM. n = 3. **, P < 0.01 (t test). (I) A model illustrating SIRT7-mediated heterochromatin stabilization in hMSCs. In young hMSCs, SIRT7 functions as a stabilizer for nuclear lamina proteins Lamin B1, LBR, and heterochromatin proteins KAP1, HP1α and HP1γ, which together form a complex at LADs to ensure chromatin spatial organization and silence LINE1. In old hMSCs, SIRT7 downregulation leads to the detachment of the complex from LADs and promotes the transcription and accumulation of LINE1 DNA, which in turn triggers activation of the cGAS-STING pathway and an immune response. 3TC is used to inhibit LINE1 reverse transcription and thus rescue senescence phenotypes
Discussion
In this study, we closed the knowledge gap between nucleolar sirtuin SIRT7 function and hMSC senescence, revealing that SIRT7 is required for the association between the nuclear lamina and heterochromatin regions and epigenetic silencing of LINE1. Below are the highlights of our study: (1) We generated SIRT7−/− hESCs and hMSCs through CRISPR/Cas9 technology. (2) The expression of SIRT7 was downregulated during aging, which in turn functioned as a driver for hMSC senescence. (3) SIRT7 maintained heterochromatin architecture via complexing with nuclear lamina proteins and heterochromatin components. (4) Depletion of SIRT7 induced LINE1 activation and triggered senescence phenotypes via activating cGAS-STING pathway in hMSCs. (5) 3TC attenuated hMSC senescence as a LINE1’s inhibitor.
SIRT7 is reported to govern a wide array of biological processes, frequently through its enzymatic activity. For instance, SIRT7 is recruited to DNA damage sites and catalyzes both deacetylation of H3K18ac and desuccinylation of H3K122succ to protect genome integrity (Li et al., 2016; Vazquez et al., 2016). Besides, SIRT7 can act as a deglutarylase of H4K91glu, the downregulation of which is tightly associated with chromatin remodeling and DNA repair (Bao et al., 2019). Intrigued by such diverse roles of SIRT7 in different species and cellular contexts, we set out to explore novel functions of SIRT7 in human stem cells. We thereby turned to CRISPR/Cas9 gene-editing to create SIRT7-null hESCs and further differentiated them into hMSCs. In line with previous work showing that hESCs are tolerant of a variety of cellular defects caused by genetic and epigenetic aberrations (Zhang et al., 2013), we found that SIRT7 was dispensable for hESC pluripotency. In contrast, SIRT7-deficient hMSCs were vulnerable to replicative stress and showed accelerated senescence and various aging defects. Nevertheless, it is interesting to note that depletion of SIRT7 in hMSCs did not result in changes in its typical histone substrates, suggesting the existence of cell type-specific or compensatory mechanisms for endogenous SIRT7. SIRT7 complexed with nuclear lamina proteins and heterochromatin proteins, which was mechanistically required for stable heterochromatin architecture at the nuclear periphery.
To our knowledge, we provide the first evidence that SIRT7 regulated hMSC senescence by modulating a heterochromatin-LINE1-cGAS-STING axis. Emerging evidence indicates that condensed heterochromatin is needed to safeguard genomic integrity, at least in part by silencing LINE1. For example, KAP1 facilitates the formation of repressive heterochromatin and initiates epigenetic silencing of LINE1 in early development stage (Castro-Diaz et al., 2014; Percharde et al., 2018). Moreover, SIRT6 was found to ribosylate KAP1 for silencing LINE1 in mouse embryonic fibroblasts (MEFs) (Van Meter et al., 2014). In our study, SIRT7 exerted a similar repressive effect on LINE1 yet through a different mechanism by which it facilitated the association of LINE1-contained heterochromatin to the nuclear lamina. Likewise, Serrano and co-workers recently observed the interplay between SIRT7 and LINE1 in MEFs (Vazquez et al., 2019). However, no involvement of SIRT7 in geroprotection and innate immune regulation was reported in their study. Our work imposes increased emphasis on the link between downstream mechanisms involving the activation of the cGAS-STING pathway and human stem cell aging. The cGAS-STING pathway is a sensor for accumulation of cytoplasmic DNA and activates the innate immune response. Our study, along with two very recent papers (De Cecco et al., 2019; Simon et al., 2019), suggests that increased LINE1 activity drives cGAS-STING pathway activation and induces cellular senescence. SIRT6, another nuclear-localized sirtuin family member, is not only capable of protecting hMSCs from oxidative injuries by coactivating transcription factor NRF2 (Pan et al., 2016) but also promotes healthy lifespan of mice through LINE1-cGAS-STING axis (Simon et al., 2019). SIRT6 and SIRT7 are both nuclear-localized and able to repress the LINE1-cGAS-STING axis to protect against cellular senescence. Consistent with the results that 3TC improves the health and cellular pathologies in Sirt6-deficient mice (Simon et al., 2019), we found that 3TC could ameliorate the premature aging phenotypes of SIRT7-deficient hMSCs. Our study extends the knowledge of sirtuins in regulating heterochromatin and downstream cGAS-STING pathway, which not only points to a role of SIRT7 loss in driving hMSC aging but also provides a plausible explanation for the emergence of other hMSC senescence-associated phenotypes including loss of heterochromatin, activation of LINE1, and induction of cGAS-STING pathway and related SASP.
Perhaps the most intriguing finding is that the downstream molecular programs of SIRT7 may be targeted with a small-molecule inhibitor to achieve the reversal of cellular aging. We show in this study that either lentiviral overexpression of SIRT7 or application of the small-molecule chemical 3TC rejuvenated aged hMSCs. Since hMSC senescence is functionally linked to various human aging syndromes (Zhang et al., 2015; Kubben et al., 2016; Deng et al., 2019; Fu et al., 2019; Ren et al., 2019), the approaches generated here (i.e., compound intervention and gene therapy) may be applicable to identify other compounds or molecular targets for the development of future therapies for age-related disease.
Materials and methods
Cell culture
WT-hESCs (Line H9, from WiCell Research) and SIRT7−/− hESCs were maintained on mitomycin C-inactivated MEFs in hESC culture medium (Cheng et al., 2019): 80% DMEM/F12 (Gibco), 20% Knockout Serum Replacement (Gibco), 2 mmol/L GlutaMAX (Gibco), 0.1 mmol/L non-essential amino acids (NEAA, Gibco), 55 μmol/L β-mercaptoethanol (Invitrogen), and 10 ng/mL bFGF (Joint Protein Central); hESCs were also cultured on Matrigel (BD Biosciences) with mTeSR medium (STEMCELL Technologies, Vancouver). All hMSCs were cultured in MSC culture medium: 90% α-MEM with GlutaMAX (Gibco), 10% fetal bovine serum (FBS, Lot A77E01F, Gemcell), 1% penicillin/streptomycin (Gibco) and 1 ng/mL bFGF (Joint Protein Central). 10 μmol/L Lamivudine (3TC) (L1295-50mg, Sigma-Aldrich) was used to treat with hMSCs.
Knockout of SIRT7 gene in WT hESCs
CRISPR/Cas9-mediated gene editing (Suzuki et al., 2016) was performed as previously described (Wang et al., 2020) with a variety of modifications. In brief, SIRT7 gRNA (CAGCGTCTATCCCAGACTAC) targeting exon 4 of SIRT7 was cloned into gRNA-mCherry vector (SIRT7-gRNA-mCherry). The pCAG-1BPNLS-Cas9-1BPNLS-2AGFP plasmid was purchased from Addgene (#87109). After electroporation, cells were seeded on Matrigel-coated plates and treated with ROCK inhibitor (Tocris) in mTeSR. After 48 h of expansion, dual-positive cells were collected by FACS (BD FACS Aria II) and plated on MEF feeder cells in hESC medium. Emerging clones were manually picked into 24-well plates and then genomic DNAs of the clones were extracted for PCR and sequencing. The corrected hESC clones were homozygous for the addition of base A, resulting in TAA premature termination at both alleles and thus leading to complete knockout of SIRT7. hESC clones were then expanded on Matrigel-coated plates and gene knockout was confirmed by Western blot analysis. Primer sequences are shown in Table S1.
Generation of hMSCs
Embryoid bodies were left to differentiate in hMSC differentiation medium (Yan et al., 2019) supplemented with MEMα Gibco, 10% FBS (Lot A77E01F, Gemcell), 10 ng/mL bFGF (Joint Protein Central), and 5 ng/mL TGFβ (Humanzyme) until fibroblast-like cells appeared. hESC-derived hMSCs were then FACS-purified with different antibodies corresponding to hMSC-specific markers (CD73, CD90, and CD105). Surface antigens CD44, CD166, CD29, HLA-ABC and CD13 were used as hMSC positive markers, and CD164, CD14, CD19 and PDPN as hMSC negative markers. The tri-lineage differentiation potential of hMSC lines was evaluated using von Kossa (osteogenesis), Alcian blue (chondrogenesis), and Oil red O kit (IHC World) (adipogenesis) (Wang et al., 2018).
Lentiviral shRNA and CRISPR/Cas9-mediated gene knockdown
STING-specific shRNAs cloned into pLVTHM plasmids (Addgene, #12247) were co-transfected with psPAX2 (Addgene, #12260) and pMD2.G (Addgene, #12259) plasmids into HEK293T cells for lentivirus production and viruses were then collected with ultracentrifugation at 19,400 ×g for 2.15 h. SIRT7-specific sgRNAs were cloned into lenti-CRISPRv2 (Addgene, #52961) with an hSpCas9 expression cassette for Lentiviral CRISPR/Cas9-mediated gene editing. Lentivirus-mediated depletion of SIRT7 was performed in WT hMSCs. For the enrichment of lentivirus-transduced cells, cells were treated with 0.5 μg/mL puromycin (Gibco) at 72 h after infection. Primer sequences are shown in Table S2.
Isolation and culture of primary hMSCs
Primary hMSCs were isolated from different individuals at the No. 306 Hospital of PLA, under the approval by the ethics committee (Zhang et al., 2015; Deng et al., 2019; Fu et al., 2019; Ren et al., 2019; Yan et al., 2019). Dental pulp tissues were processed into small pieces in TrypLE™ Express Enzyme (1×) plus Dispase IV and further digested at 37 °C for 30 min. The mixture was filtered by a cell strainer (70 μm), and then centrifuged at 500 ×g for 10 min (RT). The pellets consist of supernatant were cultivated on Gelatin-coated plates with MSC culture medium overnight. The culture medium for hMSCs was changed every other day. The sample information is shown in Table S2.
Western blot analysis
Cells were lysed in 2× SDS buffer (Sigma-Aldrich) for analysis of non-phosphorylated proteins, and in RIPA buffer for analysis of phosphorylated proteins. To measure the protein concentration, we used the BCA Kit from Thermo Fisher Scientific. Approximately 20 μg protein per sample was subjected to SDS-PAGE electrophoresis and then transferred to PVDF membranes (Millipore). After a blocking step with 5% milk (BBI Life Sciences), membranes were incubated with different primary antibodies and corresponding HRP-conjugated secondary antibodies. The quantification of image was used by Image Lab software (ChemiDoc XRS+ system, Bio-Rad).
Immunofluorescence microscopy
Cells seeded on coverslips (Thermo Fisher Scientific) were washed twice in PBS and fixed in 4% paraformaldehyde (PFA) for 30 min. 0.4% Triton X-100 (Sigma) was diluted by PBS, in which cells were permeabilized for 30 min. Then, cells were blocked in 10% donkey serum in PBS (Jackson ImmunoResearch) for 1 h (RT). After incubation with primary antibodies in blocking buffer at 4 °C overnight, cells were incubated with secondary antibodies at room temperature for 1 h and labeled with Hoechst33342 (Thermo Fisher Scientific) to visualize nuclei. Images were taken with a Leica SP5 confocal microscope.
Transmission electron microscope (TEM)
WT and SIRT7-deficient hMSCs at middle passage (P6) were harvested by TrypLE™ (Thermo Fisher Scientific) and centrifuged at 500 ×g for 5 min (RT). The collected pellets were fixed with 4% PFA in PBS, pH 7.4, on ice overnight. Using graded series of ethanol, cells were dehydrated and then infiltrated with Lowicryl resin HM20. Images were collected with a Spirit transmission electron microscope (FEI Company) operating at 100 kV.
Plasmid construction
To generate plasmids overexpressing SIRT7, KAP1, HP1α, Lamin B1 or luciferase (Luc, used as control), cDNAs were cloned into the pLE4 vector (a gift from Tomoaki Hishida) (Deng et al., 2019).
SA-β-gal staining
SA-β-gal staining was performed as previously described (Ren et al., 2019). Briefly, cultured cells were fixed for 5 min (RT) in the stationary liquid mixed by 0.2% glutaraldehyde and 2% formaldehyde. Fixed cells were stained with SA-β-gal staining solution at 37 °C overnight, percentages of SA-β-gal-positive cells were then calculated.
Clonal expansion assay
Clonal expansion assays were performed as previously described (Fu et al., 2019). Briefly, 2,000 MSCs were seeded in a Gelatin-coated 12-well plate. Harvested cells were fixed in 4% paraformaldehyde (PFA) for 30 min, and stained with 0.2% crystal violet for 1 h (RT). Relative cell density was quantified with Image J.
Measurement of ROS level
For ROS measurements, living cells were incubated with ROS indicators (1 μmol/L CM-H2DCFDA, C6827, Molecular Probes). All measurements were achieved with an LSRFortessa cell analyzer (BD), and the data were analyzed with FlowJo software (TreeStar, Ashland, OR).
Co-immunoprecipitation (co-IP)
HEK293T cells were transfected with plasmids expressing Flag-Luc (control) and Flag-SIRT7. Cells were then lysed in the lysis buffer (120 mmol/L NaCl, 0.3% CHAPS, 1 mmol/L EDTA, 40 mmol/L HEPES, pH 7.5, and complete protease inhibitor cocktail (Roche)). SIRT7+/+ hMSCs were lysed in CHAPS lysis buffer for endogenous co-IP. HEK293T cells or hMSCs were lysed at 4 °C for 2 h and then centrifuged at 12,000 ×g at 4 °C for 30 min. For endogenous co-IP, lysates (1 mg protein) were pre-cleared with 20 μL of Protein A/G-PLUS Agarose beads (Santa Cruz) for 2 h, and the supernatants were then collected by centrifugation at 3,000 rpm at 4 °C for 3 min. Next, supernatants were mixed with indicated antibodies and the beads were rotated at 4 °C overnight. The immunocomplexes were washed three times with CHAPS buffer and then eluted by boiling in 2× SDS-loading buffer for 10 min.
LC-MS/MS analysis
Protein eluates from IP were separated by 10% SDS-PAGE gel. The gel was stained with Coomassie brilliant blue and then washed with water for several times to destain. After determining molecular weight, bands were excised from the gel for aimed proteins. Samples then underwent dehydration (100% acetonitrile), reduction (10 mM DTT in 25 mmol/L NH4HCO3 for 45 min at 56 °C) and alkylation (40 mmol/L iodoacetamide in 25 mmol/L NH4HCO3 for 45 min at room temperature in the dark). After sequence-grade modified trypsin (40 ng for each band) in 25 mmol/L NH4HCO3, gel bands were dried and digested. Finally, the enzymatic reaction was stopped with formic acid. The resultant solution was identified using nanoLC-Q EXACTIVE (Thermo Scientific) equipped with data-dependent mode that allows MS data acquirement at a high resolution 70,000 (m/z 200) across the mass range of 300–1,600 m/z. Using Sequest HT and search engine, all raw files were processed from Q Exactive and analyzed with Proteome Discovery version 1.4 for protein identification and Percolator for FDR (false discovery rate). Our data were analyzed against a Uniprot human protein database (updated on 06-2013). Percolator used FDR analysis. We defined FDR <1% for protein identification. The confidence of peptides was set as high for peptides filter. Gene Ontology enrichment analysis was conducted by ToppGene (Chen et al., 2009).
Antibodies
The antibodies used for Western blot analysis are as follows: anti-SIRT7 (#5360S, 1:1,000), anti-HP1α (#2616S, 1:1,000) anti-HP1γ (#2619, 1:3,000), anti-p-TBK1 (#5483S, 1:1,000), anti-p-IRF3 (#4947S, 1:1,000), anti-p-P65 (#3033S, 1:1,000), anti-P21 (#2947S, 1:1,000) and anti-H3K36ac (07-354, 1:1,000) from Cell Signaling Technology, anti-KAP1 (Ab22553, 1:2,000), anti-LBR (Ab32535, 1:1,000), anti-Lamin B1 (Ab16048, 1:1,000), anti-LINE1-ORF2p (Ab106004, 1:500) and anti-H3K18ac (Ab1191, 1:2,000) from Abcam, anti-β-Tubulin (sc-5274, 1:3,000) and anti-H3 (sc-10809, 1:2,000) from Santa Cruz Biotechnology, anti-LAP2 (611000, 1:500) and anti-P16 (550834, 1:500) from BD Biosciences, anti-Flag (#F1804, 1:3,000) and anti-H3K36ac (07-354, 1:1,000) from Sigma-Aldrich, and anti-H3K122succ (PTM-413 1:3,000) from PTM BioLabs.
The antibodies used for immunostaining are as follows: anti-Ki67 (ZM0166, 1:500) from ZSGB-BIO, anti-phospho-Histone H2AX (Ser139) (05636, 1:400), anti-LINE1-ORF1p (MABC1152, 1:1,000) from Millipore, anti-53BP1 (A300-273A, 1:600) from Bethyl Laboratories, anti-Lamin A/C (sc-376248, 1:500), anti-OCT3/4 (sc-5279, 1:200) and anti-SOX2 (sc-17320, 1:100) from Santa Cruz Biotechnology, anti-H3K9me3 (Ab8898, 1:500) and anti-NANOG (Ab21624, 1:200) from Abcam, and anti-FABP4 (AF3150, 1:100) from R&D Systems.
The antibodies used for flow cytometry are as follows: anti-CD73 (550741, 1:200), anti-CD90 (555595, 1:200), anti-CD14 (555398, 1:200), anti-CD44 (550989, 1:200), anti-CD43, and anti-CD19 (555415, 1:200) from BD Biosciences, anti-CD105 (17-1057, 1:200) and anti-PDPN (17-9381-41, 1:200) from eBioscience, anti-CD29 (303004, 1:200), anti-CD166 (343903, 1:200) and anti-CD164 (324805, 1:200) from Biolegend.
DNA and RNA analyses
Total RNA at early (P3) and late passages (P8) was extracted in TRIzol™ (Thermo Fisher Scientific), reverse transcribed to cDNA using GoScript™ Reverse Transcription System for RT-qPCR (Promega) and subjected to genomic DNA removal using a DNA-free kit (Thermo Fisher Scientific). Genomic DNA was then purified with a DNA extraction kit (TIANGEN), and PCR was carried out with PrimeSTAR polymerase. RT-qPCR was performed using the qPCR Mix (TOYOBO) in a CFX384 Real-Time system (Bio-Rad). Primer sequences are shown in Table S1.
RNA-seq library construction and sequencing
Using the NEBNext® Poly (A) mRNA Magnetic Isolation Module, mRNA was isolated for RNA-seq. We constructed sequencing libraries using the NEBNext® Ultra™ RNA Library Prep Kit for Illumina following the manufacturer’s protocol and then sequenced libraries on Illumina HiSeq X-Ten platforms with paired-end 150-bp sequencing. Quality control and sequencing were done by Novogene Bioinformatics Technology.
RNA-seq data processing
Raw reads for RNA-seq libraries in WT and SIRT7-deficient cells were trimmed by the Trim Galore software (version 0.4.5) (https://github.com/FelixKrueger/TrimGalore). We then aligned cleaned reads to the UCSC human hg19 genome using hisat2 (version 2.0.4) (Kim et al., 2015). Then, the expression level (read count) for each gene was calculated counted with HTSeq (version 0.11.0), keeping only high-quality reads (mapping quality more than 20) (Anders et al., 2015). Fragments Per Kilobase per Million (FPKM) was calculated using stringTie (version 1.2.3) (Pertea et al., 2015). Gene set enrichment analysis (GSEA) was conducted by GSEA software (version 2.2.4) (Subramanian et al., 2005).
DamID-seq
DamID-seq was performed as described (Vogel et al., 2007) with modifications. The plasmids pLgw V5-EcoDam and pLgw EcoDam-V5-EMD were gifts from Prof. Bas van Steensel, NKI. WT MSCs and SIRT7−/− MSCs at P6 were transduced with Dam or Dam-EMD lentivirus. Cells were harvested after 72 h and the genomic DNA was isolated using DNeasy Blood & Tissue Kit (Qiagen). After DpnI digestion, adaptor ligation, DpnII digestion, PCR amplification and purification, the amplified DNA was sonicated and digested with AlwI (NEB) to remove the adaptors. The DNA libraries were constructed using a NEBNext® Ultra™ DNA Library Prep Kit and were then pooled and subjected to 150 bp paired-ends sequencing on Illumina NovaSeq sequencers.
DamID-seq data processing
For the processing of DamID-seq data, raw reads were trimmed by the Trim Galore software (version 0.4.5) (https://github.com/FelixKrueger/TrimGalore). Cleaned reads were aligned to UCSC human hg19 genome using Bowtie 2 (version 2.2.9) (Langmead and Salzberg, 2012). Duplicated reads were filtered out by MarkDuplicates.jar program in Picard tools (https://broadinstitute.github.io/picard/). To minimize the bias of sequencing depth, replicates for each sample were merged. Then, 120 million high-quality reads for Dam or Dam-EMD data set in WT or SIRT7−/− hMSCs were randomly selected and retained for downstream analysis. To visualize the DamID signal, we calculated the log2 transformed Reads Per Kilobase per Million mapped reads (RPKM) values of Dam-EMD and Dam signal [log2(Dam-EMD/Dam)] in WT or SIRT7−/− hMSCs for each 10-bp bin using bamCompare program deepTools (version 2.5.4-2-5ee467f) software.
To identify LAD regions in WT or SIRT7−/− hMSCs, we calculated log2 transformed RPKM values of Dam-EMD and Dam signal [log2 (Dam-EMD/Dam)] in WT or SIRT7−/− hMSCs for each 2-kb bin. We then identified LAD regions with the R package HMMt (https://github.com/dinovski/asDamID/blob/master/scripts/hmmt_functions.R).
ChIP-seq and ChIP-qPCR
ChIP-seq and ChIP-qPCR were performed following a previously reported protocol with minor modifications (Deng et al., 2019). Briefly, WT and SIRT7−/− hMSCs at middle passage (P6) were cross-linked in 1% formaldehyde at room temperature with continuous rotation for 10 min. Next, cross-linked cells were quenched with 125 mmol/L Glycine for 5 min and then lysed on ice for 10 min, followed by chromatin shearing to a target peak size of 100–500 bp using the Covaris S220 focused-ultrasonicator (Covaris). The supernatant retained after centrifugation was then incubated with 2.4 μg H3K9me3 antibody (Ab8898, Abcam) or SIRT7 antibody (#5360S, Cell Signaling Technology) pre-conjugated with Dynabeads Protein A (10002D, Thermo Fisher Scientific) overnight at 4 °C. Normal rabbit IgG (#2729S, Cell Signaling Technology) was used as a negative control. After elution and reverse crosslinking at 68 °C, fragmented DNA was purified with phenol-chloroform-isoamyl alcohol extraction and ethanol precipitation and subjected to sequencing or RT-qPCR analysis. Libraries were prepared using KAPA Hyper Prep Kits (KK8504, KAPA Biosystems) and Next Multiplex Oligos for Illumina (Index Primers Set 1) (E7335L, New England Biolabs), and then sequenced on HiSeq X-Ten platforms according to the manufacturer’s instruction. Primer sequences are shown in Table S1.
ChIP-seq data processing
Raw reads were trimmed using the Trim Galore software (version 0.4.5) (https://github.com/FelixKrueger/TrimGalore) and clean reads aligned to the UCSC human hg19 genome using Bowtie 2 (version 2.2.9) (Langmead and Salzberg, 2012). To minimize the bias of sequencing depth, replicates for each sample were merged. Then, 60 million high-quality reads were randomly selected and retained for downstream analysis. We then called H3K9me3 peaks by SICER (version 1.1) with the parameter “-w 200 -g 3” (Zang et al., 2009). H3K9me3 peaks with FDR (False discovery rate) of 1% or better were retained.
To identify “H3K9me3 mountains”, H3K9me3 signal (CPM, Count Per Million) for each H3K9me3 peak in WT or SIRT7−/− hMSCs was calculated. We then ranked H3K9me3 peaks by increasing H3K9me3 signal in WT or SIRT7−/− hMSCs as shown in Fig. S4E. These plots showed inflection points where the H3K9me3 signal began increasing rapidly. H3K9me3 peaks above the inflection point were then named as “H3K9me3 mountains”.
ATAC-seq library preparation and sequencing
ATAC-seq libraries of hMSCs were prepared as previously described (Buenrostro et al., 2013; Wu et al., 2016; Wang et al., 2019b) with some modifications. In brief, 50,000 cells of WT or SIRT7−/− hMSCs at middle passage (P6) were washed twice in cold PBS and lysed in 50 μL ice-cold lysis buffer (10 mmol/L Tris-HCl (pH 7.4), 10 mmol/L NaCl, 3 mmol/L MgCl2 and 0.5% NP-40) for 10 min. Lysates were briefly spun at 500 ×g for 5 min to obtain the nuclei. The nuclei were then incubated with the Tn5 transposome and tagmentation buffer at 37 °C for 30 min (TD501, Vazyme Biotech). After tagmentation, fragmented DNA was purified with 100 μL (2×) AMPure XP beads (A63882, Beckman Coulter) and amplified for 13 cycles using the following PCR conditions: 72 °C for 3 min; 98 °C for 30 s; and thermocycling at 98 °C for 15 s, 60 °C for 30 s and 72 °C for 30 s; following by 72 °C 5 min. PCR reaction enzyme and index primers were purchased from Vazyme Biotech (TD202). After PCR amplification, libraries were purified and selected by using 0.5× /1.3× AMPure XP beads. Finally, libraries were sequenced on HiSeq X-Ten platforms according to the manufacturer’s instruction.
ATAC-seq data processing
Raw reads were cleaned with the Trim Galore software (version 0.4.5) and trimmed reads aligned to the UCSC human hg19 genome using Bowtie 2 (version 2.2.9) with the parameter “ -X 2,000 -N 1 -L 25 –no-mixed –no-discordant -t ” (Langmead and Salzberg, 2012). Reads of PCR duplicates were removed with the MarkDuplicates.jar program in Picard tools. Replicates for each sample were merged for downstream analysis. For ATAC peak calling, MACS2 (version 2.1.1) was used with the parameter “–nomodel –shift 0 –extsize 250 –call-summits” (Zhang et al., 2008).
Whole genome sequencing and copy number variation analysis
Genomic DNA from 1 × 106 WT or SIRT7−/− hMSCs at middle passage (P6) as per duplicate was isolated using DNeasy Blood & Tissue Kit (Qiagen). Quality control and sequencing were performed following standard protocols from Novogene Bioinformatics Technology Co. Ltd. Genome-wide copy number variation (CNV) analysis was conducted as previously described (Deng et al., 2019). Raw reads were trimmed by the TrimGalore software (version 0.4.5) and clean reads aligned to the UCSC hg19 human genome using bowtie2 software (version 2.2.9). R package HMMcopy (version 1.25.0) was implemented to calculate CNVs in each 0.5 Mb bin size (Ha et al., 2012).
Assessment of the reproducibility of sequencing data
To evaluate the reproducibility of H3K9me3 ChIP-seq and ATAC-seq data sets, we first calculated RPKM level for each 2-kb bin genome widely. We then calculated the Euclidean distance by R (version 3.5.1) to evaluate reproducibility. Lower value of Euclidean distance means highly correlated. To evaluate the reproducibility of DamID-seq, principle component analysis (PCA) was calculated by R (version 3.5.1). Pearson correlation coefficient (R) between replicates was calculated based on the expression level (regularized-logarithm normalized read count) to evaluate the reproducibility of RNA-seq data.
Retrotransposition assay
WT and SIRT7−/− hMSCs were transfected with 2 μg p99-GFP-LRE3-Cherry (a gift from Alysson R. Muotri) and EGFP-positive cells were sorted by FACS at three days post-transfection. We calculated the adjusted retrotransposition rate by number of EGFP-positive cells normalized by the transfection efficiency, which was determined by the mCherry reporter, and by transfected cell numbers.
ELISA
Protein levels of IL6 and MCP1 were measured by sandwich enzyme-linked immunoassay (ELISA). Supernatants from WT and SIRT7−/− hMSCs at middle passage (P6) were collected and centrifuged at 500 ×g for 5 min to remove debris. ELISA kits from systems were used: IL6 (BioLegend) and MCP1 (R&D Systems). Plates were scanned at 450 nm using Synergy H1 (BioTek) and finally data were normalized by cell numbers.
Comet assay
Comet assay was performed as described previously (Olive and Banath, 2006) with a minor modification. The images were acquired by CKX41 microscope and then quantified using CASP software.
Animal experiments
hMSC implantation assays were performed as previously described (Fu et al., 2019). One million SIRT7+/+ or SIRT7−/− hMSCs expressing luciferase were injected into the TA muscle of male nude mice aged 6-8 weeks. In vivo luciferase activity was measured by an IVIS Spectrum imaging system (XENOGEN, Caliper).
Statistical analysis
All data in our study were presented as the means ± SEM. We used GraphPad Prism 7 Software to conduct two tailed Student’s t-test. P value < 0.05 was considered statistically significant.
Abbreviations
- 3TC:
-
Lamivudine
- ATAC-seq:
-
Assay for transposase accessible chromatin sequencing
- cGAS-STING:
-
The cyclic GMP-AMP synthase-stimulator of interferon genes
- CNV:
-
Genome-wide copy number variation
- co-IP:
-
Co-immunoprecipitation
- CRISPR/Cas9:
-
Clustered regularly interspaced short palindromic repeat/CRISPR associated gene 9
- DamID-seq:
-
DNA adenine methyltransferase identification with high-throughput sequencing
- DSBs:
-
DNA double-strand breaks
- DDR:
-
DNA damage response
- H3K9me3:
-
Histone H3 Lys9 trimethylation
- HDACs:
-
Histone deacetylases
- hESC:
-
Human embryonic stem cell
- HGPS:
-
Hutchinson-Gilford progeria syndrome
- hMSC:
-
Mesenchymal stem cell
- LC-MS/MS:
-
Liquid chromatography-tandem mass spectrometry
- LADs:
-
Lamina-associated domains
- LINE1:
-
Long interspersed element 1
- Luc:
-
Luciferase
- MEF:
-
Mouse embryonic fibroblast
- NAD:
-
Nicotinamide adenine dinucleotide
- PDPN:
-
Podoplanin
- RTis:
-
Reverse-transcriptase inhibitors
- ROS:
-
Reactive oxygen species
- RS:
-
Replicatively senescent
- SA-β-gal:
-
Senescence-associated-β-galactosidase
- SASP:
-
Senescence-associated secretory phenotype
- TA:
-
Tibialis anterior
- TE:
-
Transposable element
- WT:
-
Wild-type
References
Anders S, Pyl PT, Huber W (2015) HTSeq—a Python framework to work with high-throughput sequencing data. Bioinformatics (Oxford, England) 31:166–169
Araki S, Izumiya Y, Rokutanda T, Ianni A, Hanatani S, Kimura Y, Onoue Y, Senokuchi T, Yoshizawa T, Yasuda O et al (2015) Sirt7 contributes to myocardial tissue repair by maintaining transforming growth factor-beta signaling pathway. Circulation 132:1081–1093
Bao X, Liu Z, Zhang W, Gladysz K, Fung YME, Tian G, Xiong Y, Wong JWH, Yuen KWY, Li XD (2019) Glutarylation of Histone H4 Lysine 91 regulates chromatin dynamics. Mol Cell 76(660–675):e669
Barber MF, Michishita-Kioi E, Xi Y, Tasselli L, Kioi M, Moqtaderi Z, Tennen RI, Paredes S, Young NL, Chen K et al (2012) SIRT7 links H3K18 deacetylation to maintenance of oncogenic transformation. Nature 487:114–118
Bickmore WA, van Steensel B (2013) Genome architecture: domain organization of interphase chromosomes. Cell 152:1270–1284
Bishop NA, Guarente L (2007) Genetic links between diet and lifespan: shared mechanisms from yeast to humans. Nat Rev Genet 8:835–844
Bourque G, Burns KH, Gehring M, Gorbunova V, Seluanov A, Hammell M, Imbeault M, Izsvak Z, Levin HL, Macfarlan TS et al (2018) Ten things you should know about transposable elements. Genome Biol 19:199
Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ (2013) Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods 10:1213–1218
Castro-Diaz N, Ecco G, Coluccio A, Kapopoulou A, Yazdanpanah B, Friedli M, Duc J, Jang SM, Turelli P, Trono D (2014) Evolutionally dynamic L1 regulation in embryonic stem cells. Genes Dev 28:1397–1409
Chen J, Bardes EE, Aronow BJ, Jegga AG (2009) ToppGene Suite for gene list enrichment analysis and candidate gene prioritization. Nucl Acids Res 37:W305–311
Chen S, Blank MF, Iyer A, Huang B, Wang L, Grummt I, Voit R (2016) SIRT7-dependent deacetylation of the U3-55k protein controls pre-rRNA processing. Nat Commun 7:10734
Cheng F, Wang S, Song M, Liu Z, Liu P, Wang L, Wang Y, Zhao Q, Yan K, Chan P et al (2019) DJ-1 is dispensable for human stem cell homeostasis. Protein Cell 10:846–853
Cioffi M, Vallespinos-Serrano M, Trabulo SM, Fernandez-Marcos PJ, Firment AN, Vazquez BN, Vieira CR, Mulero F, Camara JA, Cronin UP et al (2015) MiR-93 controls adiposity via inhibition of Sirt7 and Tbx3. Cell Rep 12:1594–1605
Dai L, Huang Q, Boeke JD (2011) Effect of reverse transcriptase inhibitors on LINE-1 and Ty1 reverse transcriptase activities and on LINE-1 retrotransposition. BMC Biochem 12:18
De Cecco M, Criscione SW, Peckham EJ, Hillenmeyer S, Hamm EA, Manivannan J, Peterson AL, Kreiling JA, Neretti N, Sedivy JM (2013) Genomes of replicatively senescent cells undergo global epigenetic changes leading to gene silencing and activation of transposable elements. Aging Cell 12:247–256
De Cecco M, Ito T, Petrashen AP, Elias AE, Skvir NJ, Criscione SW, Caligiana A, Brocculi G, Adney EM, Boeke JD et al (2019) L1 drives IFN in senescent cells and promotes age-associated inflammation. Nature 566:73–78
Deng L, Ren R, Liu Z, Song M, Li J, Wu Z, Ren X, Fu L, Li W, Zhang W et al (2019) Stabilizing heterochromatin by DGCR8 alleviates senescence and osteoarthritis. Nat Commun 10:3329
Dimarino AM, Caplan AI, Bonfield TL (2013) Mesenchymal stem cells in tissue repair. Front Immunol 4:201
Finkel T, Deng CX, Mostoslavsky R (2009) Recent progress in the biology and physiology of sirtuins. Nature 460:587–591
Fu L, Hu Y, Song M, Liu Z, Zhang W, Yu FX, Wu J, Wang S, Izpisua Belmonte JC, Chan P et al (2019) Up-regulation of FOXD1 by YAP alleviates senescence and osteoarthritis. PLoS Biol 17:e3000201
Garcia-Perez JL, Morell M, Scheys JO, Kulpa DA, Morell S, Carter CC, Hammer GD, Collins KL, O’Shea KS, Menendez P et al (2010) Epigenetic silencing of engineered L1 retrotransposition events in human embryonic carcinoma cells. Nature 466:769–773
Geng L, Liu Z, Wang S, Sun S, Ma S, Liu X, Chan P, Sun L, Song M, Zhang W et al (2019) Low-dose quercetin positively regulates mouse healthspan. Protein Cell 10:770–775
Gorbunova V, Boeke JD, Helfand SL, Sedivy JM (2014) Human genomics. Sleeping dogs of the genome. Science 346:1187–1188
Grewal SI, Jia S (2007) Heterochromatin revisited. Nature reviews. Genetics 8:35–46
Guelen L, Pagie L, Brasset E, Meuleman W, Faza MB, Talhout W, Eussen BH, de Klein A, Wessels L, de Laat W et al (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453:948–951
Ha G, Roth A, Lai D, Bashashati A, Ding J, Goya R, Giuliany R, Rosner J, Oloumi A, Shumansky K (2012) Integrative analysis of genome-wide loss of heterozygosity and monoallelic expression at nucleotide resolution reveals disrupted pathways in triple-negative breast cancer. Genome Res 22:1995–2007
He X, Memczak S, Qu J, Belmonte JCI, Liu G-H (2020) Single-cell omics in ageing: a young and growing field. Nat Metab 2:293–302
Herskovits AZ, Guarente L (2013) Sirtuin deacetylases in neurodegenerative diseases of aging. Cell Res 23:746–758
Iyer-Bierhoff A, Krogh N, Tessarz P, Ruppert T, Nielsen H, Grummt I (2018) SIRT7-dependent deacetylation of fibrillarin controls histone H2A methylation and rRNA synthesis during the cell cycle. Cell Rep 25(2946–2954):e2945
Jones RB, Garrison KE, Wong JC, Duan EH, Nixon DF, Ostrowski MA (2008) Nucleoside analogue reverse transcriptase inhibitors differentially inhibit human LINE-1 retrotransposition. PLoS ONE 3:e1547
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357–360
Kiran S, Oddi V, Ramakrishna G (2015) Sirtuin 7 promotes cellular survival following genomic stress by attenuation of DNA damage, SAPK activation and p53 response. Exp Cell Res 331:123–141
Kubben N, Misteli T (2017) Shared molecular and cellular mechanisms of premature ageing and ageing-associated diseases. Nat Rev Mol Cell Biol 18:595–609
Kubben N, Zhang W, Wang L, Voss TC, Yang J, Qu J, Liu GH, Misteli T (2016) Repression of the antioxidant NRF2 pathway in premature aging. Cell 165:1361–1374
Kudlow BA, Kennedy BK, Monnat RJ Jr (2007) Werner and Hutchinson-Gilford progeria syndromes: mechanistic basis of human progeroid diseases. Nat Rev Mol Cell Biol 8:394–404
Langmead B, Salzberg SL (2012) Fast gapped-read alignment with Bowtie 2. Nat Methods 9:357–359
Li L, Shi L, Yang S, Yan R, Zhang D, Yang J, He L, Li W, Yi X, Sun L et al (2016) SIRT7 is a histone desuccinylase that functionally links to chromatin compaction and genome stability. Nat Commun 7:12235
Liu GH, Barkho BZ, Ruiz S, Diep D, Qu J, Yang SL, Panopoulos AD, Suzuki K, Kurian L, Walsh C et al (2011) Recapitulation of premature ageing with iPSCs from Hutchinson-Gilford progeria syndrome. Nature 472:221–225
Liu GH, Qu J, Suzuki K, Nivet E, Li M, Montserrat N, Yi F, Xu X, Ruiz S, Zhang W et al (2012) Progressive degeneration of human neural stem cells caused by pathogenic LRRK2. Nature 491:603–607
Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G (2013) The hallmarks of aging. Cell 153:1194–1217
Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I (2005) Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell 16:4623–4635
Miura Y (2016) Human bone marrow mesenchymal stromal/stem cells: current clinical applications and potential for hematology. Int J Hematol 103:122–128
Mohrin M, Shin J, Liu Y, Brown K, Luo H, Xi Y, Haynes CM, Chen D (2015) Stem cell aging. A mitochondrial UPR-mediated metabolic checkpoint regulates hematopoietic stem cell aging. Science 347:1374–1377
Obeid M, Saber Sel D, Ismael Ael D, Hassanien E (2013) Mesenchymal stem cells promote hard-tissue repair after direct pulp capping. J Endod 39:626–631
Olive PL, Banath JP (2006) The comet assay: a method to measure DNA damage in individual cells. Nat Protoc 1:23–29
Pan H, Guan D, Liu X, Li J, Wang L, Wu J, Zhou J, Zhang W, Ren R, Zhang W et al (2016) SIRT6 safeguards human mesenchymal stem cells from oxidative stress by coactivating NRF2. Cell Res 26:190–205
Paredes S, Angulo-Ibanez M, Tasselli L, Carlson SM, Zheng W, Li TM, Chua KF (2018) The epigenetic regulator SIRT7 guards against mammalian cellular senescence induced by ribosomal DNA instability. The Journal of biological chemistry 293:11242–11250
Percharde M, Lin CJ, Yin Y, Guan J, Peixoto GA, Bulut-Karslioglu A, Biechele S, Huang B, Shen X, Ramalho-Santos M (2018) A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 174(391–405):e319
Pertea M, Pertea GM, Antonescu CM, Chang TC, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33:290–295
Ren R, Ocampo A, Liu GH, Izpisua Belmonte JC (2017) Regulation of stem cell aging by metabolism and epigenetics. Cell Metab 26:460–474
Ren X, Hu B, Song M, Ding Z, Dang Y, Liu Z, Zhang W, Ji Q, Ren R, Ding J et al (2019) Maintenance of nucleolar homeostasis by CBX4 alleviates senescence and osteoarthritis. Cell Rep 26(3643–3656):e3647
Sanjana NE, Shalem O, Zhang F (2014) Improved vectors and genome-wide libraries for CRISPR screening. Nat Methods 11:783–784
Secunda R, Vennila R, Mohanashankar AM, Rajasundari M, Jeswanth S, Surendran R (2015) Isolation, expansion and characterisation of mesenchymal stem cells from human bone marrow, adipose tissue, umbilical cord blood and matrix: a comparative study. Cytotechnology 67:793–807
Shin J, He M, Liu Y, Paredes S, Villanova L, Brown K, Qiu X, Nabavi N, Mohrin M, Wojnoonski K et al (2013) SIRT7 represses Myc activity to suppress ER stress and prevent fatty liver disease. Cell Rep 5:654–665
Simon M, Van Meter M, Ablaeva J, Ke Z, Gonzalez RS, Taguchi T, De Cecco M, Leonova KI, Kogan V, Helfand SL et al (2019) LINE1 derepression in aged wild-type and SIRT6-deficient mice drives inflammation. Cell Metab 29(871–885):e875
Singh PP, Demmitt BA, Nath RD, Brunet A (2019) The genetics of aging: a vertebrate perspective. Cell 177:200–220
Sridharan R, Gonzales-Cope M, Chronis C, Bonora G, McKee R, Huang C, Patel S, Lopez D, Mishra N, Pellegrini M et al (2013) Proteomic and genomic approaches reveal critical functions of H3K9 methylation and heterochromatin protein-1gamma in reprogramming to pluripotency. Nat Cell Biol 15:872–882
Stenderup K, Justesen J, Clausen C, Kassem M (2003) Aging is associated with decreased maximal life span and accelerated senescence of bone marrow stromal cells. Bone 33:919–926
Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES et al (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550
Suzuki K, Tsunekawa Y, Hernandez-Benitez R, Wu J, Zhu J, Kim EJ, Hatanaka F, Yamamoto M, Araoka T, Li Z et al (2016) In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540:144–149
Talens RP, Christensen K, Putter H, Willemsen G, Christiansen L, Kremer D, Suchiman HE, Slagboom PE, Boomsma DI, Heijmans BT (2012) Epigenetic variation during the adult lifespan: cross-sectional and longitudinal data on monozygotic twin pairs. Aging Cell 11:694–703
Thomas CA, Tejwani L, Trujillo CA, Negraes PD, Herai RH, Mesci P, Macia A, Crow YJ, Muotri AR (2017) Modeling of TREX1-Dependent autoimmune disease using human stem cells highlights L1 accumulation as a source of neuroinflammation. Cell Stem Cell 21(319–331):e318
Tsai YC, Greco TM, Cristea IM (2014) Sirtuin 7 plays a role in ribosome biogenesis and protein synthesis. Mol Cell Proteomics MCP 13:73–83
Uccelli A, Moretta L, Pistoia V (2008) Mesenchymal stem cells in health and disease. Nat Rev Immunol 8:726–736
Vakhrusheva O, Smolka C, Gajawada P, Kostin S, Boettger T, Kubin T, Braun T, Bober E (2008) Sirt7 increases stress resistance of cardiomyocytes and prevents apoptosis and inflammatory cardiomyopathy in mice. Circ Res 102:703–710
Van Meter M, Kashyap M, Rezazadeh S, Geneva AJ, Morello TD, Seluanov A, Gorbunova V (2014) SIRT6 represses LINE1 retrotransposons by ribosylating KAP1 but this repression fails with stress and age. Nat Commun 5:5011
van Steensel B, Dekker J (2010) Genomics tools for unraveling chromosome architecture. Nat Biotechnol 28:1089–1095
Vazquez BN, Thackray JK, Simonet NG, Kane-Goldsmith N, Martinez-Redondo P, Nguyen T, Bunting S, Vaquero A, Tischfield JA, Serrano L (2016) SIRT7 promotes genome integrity and modulates non-homologous end joining DNA repair. EMBO J 35:1488–1503
Vazquez BN, Thackray JK, Simonet NG, Chahar S, Kane-Goldsmith N, Newkirk SJ, Lee S, Xing J, Verzi MP, An W et al (2019) SIRT7 mediates L1 elements transcriptional repression and their association with the nuclear lamina. Nucl Acids Res 47:7870–7885
Vogel MJ, Peric-Hupkes D, van Steensel B (2007) Detection of in vivo protein-DNA interactions using DamID in mammalian cells. Nat Protoc 2:1467–1478
Volkman HE, Stetson DB (2014) The enemy within: endogenous retroelements and autoimmune disease. Nat Immunol 15:415–422
Wang P, Liu Z, Zhang X, Li J, Sun L, Ju Z, Li J, Chan P, Liu GH, Zhang W et al (2018) CRISPR/Cas9-mediated gene knockout reveals a guardian role of NF-kappaB/RelA in maintaining the homeostasis of human vascular cells. Protein Cell 9:945–965
Wang WW, Angulo-Ibanez M, Lyu J, Kurra Y, Tong Z, Wu B, Zhang L, Sharma V, Zhou J, Lin H et al (2019a) A click chemistry approach reveals the chromatin-dependent histone H3K36 deacylase nature of SIRT7. J Am Chem Soc 141:2462–2473
Wang Y, Lu T, Sun G, Zheng Y, Yang S, Zhang H, Hao S, Liu Y, Ma S, Zhang H et al (2019b) Targeting of apoptosis gene loci by reprogramming factors leads to selective eradication of leukemia cells. Nat Commun 10:5594
Wang S, Min Z, Ji Q, Geng L, Su Y, Liu Z, Hu H, Wang L, Zhang W, Suzuiki K et al (2020) Rescue of premature aging defects in Cockayne syndrome stem cells by CRISPR/Cas9-mediated gene correction. Protein Cell 11:1–22
Wronska A, Lawniczak A, Wierzbicki PM, Kmiec Z (2016) Age-related changes in sirtuin 7 expression in calorie-restricted and refed rats. Gerontology 62:304–310
Wu J, Huang B, Chen H, Yin Q, Liu Y, Xiang Y, Zhang B, Liu B, Wang Q, Xia W et al (2016) The landscape of accessible chromatin in mammalian preimplantation embryos. Nature 534:652–657
Wu Z, Zhang W, Song M, Wang W, Wei G, Li W, Lei J, Huang Y, Sang Y, Chan P et al (2018) Differential stem cell aging kinetics in Hutchinson-Gilford progeria syndrome and Werner syndrome. Protein Cell 9:333–350
Yan P, Li Q, Wang L, Lu P, Suzuki K, Liu Z, Lei J, Li W, He X, Wang S et al (2019) FOXO3-engineered human ESC-derived vascular cells promote vascular protection and regeneration. Cell Stem Cell 24(447–461):e448
Yang YK (2018) Aging of mesenchymal stem cells: implication in regenerative medicine. Regen Ther 9:120–122
Yoshizawa T, Karim MF, Sato Y, Senokuchi T, Miyata K, Fukuda T, Go C, Tasaki M, Uchimura K, Kadomatsu T et al (2014) SIRT7 controls hepatic lipid metabolism by regulating the ubiquitin-proteasome pathway. Cell Metab 19:712–721
Zang C, Schones DE, Zeng C, Cui K, Zhao K, Peng W (2009) A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics (Oxford, England) 25:1952–1958
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, Nusbaum C, Myers RM, Brown M, Li W et al (2008) Model-based analysis of ChIP-Seq (MACS). Genome Biol 9:R137
Zhang J, Lian Q, Zhu G, Zhou F, Sui L, Tan C, Mutalif RA, Navasankari R, Zhang Y, Tse HF et al (2011) A human iPSC model of Hutchinson Gilford Progeria reveals vascular smooth muscle and mesenchymal stem cell defects. Cell Stem Cell 8:31–45
Zhang W, Qu J, Suzuki K, Liu GH, Izpisua Belmonte JC (2013) Concealing cellular defects in pluripotent stem cells. Trends Cell Biol 23:587–592
Zhang W, Li J, Suzuki K, Qu J, Wang P, Zhou J, Liu X, Ren R, Xu X, Ocampo A et al (2015) Aging stem cells. A Werner syndrome stem cell model unveils heterochromatin alterations as a driver of human aging. Science 348:1160–1163
Zhang W, Wan H, Feng G, Qu J, Wang J, Jing Y, Ren R, Liu Z, Zhang L, Chen Z et al (2018) SIRT6 deficiency results in developmental retardation in cynomolgus monkeys. Nature 560:661–665
Zhang, W., Qu, J., Liu, G.H., and Belmonte, J.C.I. (2020). The ageing epigenome and its rejuvenation. Nature reviews Molecular cell biology.
Zhou S, Greenberger JS, Epperly MW, Goff JP, Adler C, Leboff MS, Glowacki J (2008) Age-related intrinsic changes in human bone-marrow-derived mesenchymal stem cells and their differentiation to osteoblasts. Aging Cell 7:335–343
Data availability
High-throughput sequencing data generated in this study have been deposited in Gene Expression Omnibus (GEO) database with accession number GSE146247.
Acknowledgments
We would like to express our gratitude to Dr. Bas van Steensel for pLgw V5-EcoDam and pLgw EcoDam-V5-EMD plasmids and Dr. Alysson R. Muotri for p99-GFP-LRE3-Cherry plasmid. We thank J. Jia (IBP, CAS) and S. Sun (IBP, CAS) for their help on FACS experiments, J. Wang (IBP, CAS) for liquid chromatography-tandem mass spectrometry (LC-MS/MS), C. Peng (IBP, CAS) for TEM sample preparation, R. Wang for gene-editing experiments, W. Li, J. Jia, H. Zhao and J. Lei for animal experiments, C. Liang, H. Hu, Y. Zhang and M. Li for cell experiments, Q. Chu, L. Bai, R. Bai, J. Lu, X. Zhuo, S. Ma and Y. Yang for administrative assistance. This work was supported by the National Key Research and Development Program of China (2017YFA0103304), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA16010100), the National Key Research and Development Program of China (2018YFC2000100, 2018YFA0107203, 2017YFA0102802), the National Natural Science Foundation of China (81625009, 91749202, 81861168034, 81921006, 31671429, 91949209, 91749123, 81671377, 81822018, 81870228, 81922027, 81701388, 31900524), the Program of the Beijing Municipal Science and Technology Commission (Z191100001519005), Beijing Natural Science Foundation (Z190019), Beijing Municipal Commission of Health and Family Planning (PXM2018_026283_000002), Advanced Innovation Center for Human Brain Protection (3500-1192012), the Key Research Program of the Chinese Academy of Sciences (KFZD-SW-221), K.C. Wong Education Foundation (GJTD-2019-06, GJTD-2019-08), Young Elite Scientists Sponsorship Program by CAST, Youth Innovation Promotion Association of CAS, the State Key Laboratory of Stem Cell and Reproductive Biology and the State Key Laboratory of Membrane Biology.
Compliance with Ethics Guidelines
The authors declare no conflict of interest.
Author information
Authors and Affiliations
Corresponding authors
Additional information
Shijia Bi and Zunpeng Liu have contributed equally to this paper as the first authors.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Bi, S., Liu, Z., Wu, Z. et al. SIRT7 antagonizes human stem cell aging as a heterochromatin stabilizer. Protein Cell 11, 483–504 (2020). https://doi.org/10.1007/s13238-020-00728-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13238-020-00728-4