Abstract
The development of gastritis is associated with an increased risk of gastric cancer. Current invasive gastritis diagnostic methods are not suitable for monitoring progress. In this work based on 78 gastritis patients and 50 healthy individuals, we observed that the variation of tongue-coating microbiota was associated with the occurrence and development of gastritis. Twenty-one microbial species were identified for differentiating tongue-coating microbiomes of gastritis and healthy individuals. Pathways such as microbial metabolism in diverse environments, biosynthesis of antibiotics and bacterial chemotaxis were up-regulated in gastritis patients. The abundance of Campylobacter concisus was found associated with the gastric precancerous cascade. Furthermore, Campylobacter concisus could be detected in tongue coating and gastric fluid in a validation cohort containing 38 gastritis patients. These observations provided biological evidence of tongue diagnosis in traditional Chinese medicine, and indicated that tongue-coating microbiome could be a potential non-invasive biomarker, which might be suitable for long-term monitoring of gastritis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gastritis, which is a worldwide problem, is defined as an inflamed condition of the gastric mucosa (Price, 1991; Stolte and Meining, 2001; Owen, 2003; Rugge et al., 2007). Gastritis development is a multistep and multifactorial process (Guo et al., 2017). The stage of gastritis can be divided into superficial gastritis, atrophic gastritis, intestinal metaplasia and dysplasia (Correa, 1992; Correa and Piazuelo, 2012). Long-term studies have confirmed that the development of gastritis increases the risk of gastric cancer (Sipponen et al., 1985; Filipe et al., 1994; Miehlke et al., 1998; Meining et al., 2002; Ohata et al., 2004; Song et al., 2015). Gastritis diagnosis in clinical practice relies primarily on endoscopy and histological examination (Dixon et al., 1996; Rugge et al., 2007), which are invasive procedures that cannot be done frequently. Hence, monitoring and controlling at regular intervals with non-invasive methods are highly demanded in the prevention and treatment of gastritis. Therefore, it is important to find biomarkers associated with the occurrence and development of gastritis.
Bacteria were shown to contribute to gastritis when Barry Marshall and Robin Warren found in 1984 that Helicobacter pylori (HP) in the stomach played an important role in chronic gastritis (Marshall and Warren, 1984). However, it was reported that approximately 25% of chronic gastritis patients were not infected by Helicobacter pylori, suggesting that some other bacteria or factors, which are capable of causing inflammation, are involved (Jonkers et al., 1997). Since then, a series of studies based on different cohorts in stomach have confirmed the role of bacteria other than Helicobacter pylori in gastric lesions (Sjostedt et al., 1985; Sahay et al., 1995; Li et al., 2009; Hakalehto et al., 2011; Aviles-Jimenez et al., 2014; Eun et al., 2014; Schulz et al., 2016; Ferreira et al., 2017; Sohn et al., 2017). The Firmicutes phylum and the Streptococcus genus were found to exhibit increased abundance in gastritis patients compared to normal controls (Li et al., 2009). Analyses of gastric mucosa microbial changes of superficial gastritis (SG), atrophic gastritis (AG), intestinal metaplasia (IM) and gastric cancer (GC) found that gastric microbial composition and interaction shifted, indicating that gastritis development is associated with shifting human microbiota (Aviles-Jimenez et al., 2014; Eun et al., 2014; Coker et al., 2017).
Among diseases in the digestive tract, gastritis status may be reflected in the coating of tongue, which is the initial part of the digestive tract. On the one hand, food and microbes transferred into the stomach could remain in the residue on the tongue coating. On the other hand, the gastroesophageal reflux of gastritis patients could bring materials from the stomach to the tongue coating. The association between difference in tongue coating microbiota and the occurrence of gastritis has been explored in several studies. Our previous work focused on the tongue coating of gastritis patients and initially found 123 and 258 species-level OTUs enriched in the tongue coating of gastritis with Cold Syndrome and that with Hot Syndrome, respectively. Moreover, tongue-coating images of healthy controls and gastritis patients were distinguishable (Jiang et al., 2012). Sun and colleagues confirmed that microbial components of tongue coating in chronic gastritis patients were different from those in normal controls based on 16S ribosomal RNA denatured gradient gel electrophoresis (Sun et al., 2013). Ye et al. found that Bacillus was present only in the yellow tongue coating of chronic erosive gastritis patients by using Illumina Miseq sequencing of the V4–V5 region of the 16S ribosomal RNA gene (Ye et al., 2016). Furthermore, for traditional Chinese medicine (TCM), tongue appearance is a major indicator of physical status including that of the stomach. Our previous work also confirmed that tongue images could be used to classify gastritis patients and healthy volunteers and to classify gastritis patients with different TCM subtypes (Kanawong et al., 2012). However, as far as we know, there are no systematical analyses based on metagenomic sequencing to reveal the tongue coating microbiome variation associated with the occurrence and development of gastritis. This study focused on whether the gastric condition is reflected in the tongue-coating microbiome, which might be objective, non-invasive and suitable for long-term monitoring.
In this study, we collected the tongue coating samples of 78 gastritis patients and 50 healthy subjects. Metagenomic analysis revealed that the variation in tongue-coating microbiota was associated with the occurrence and development of gastritis. A network including 21 tongue-coating species that differentiated the tongue-coating microbiomes of gastritis patients and healthy controls was identified. Pathways such as microbial metabolism in diverse environments, biosynthesis of antibiotics and bacterial chemotaxis were up-regulated in gastritis patients. Furthermore, the abundance of Campylobacter concisus was detected to be associated with the precancerous cascade of gastritis. To test whether Campylobacter concisus existed in the stomach, we detected the Campylobacter concisus in tongue coating and gastric fluid in a validation cohort of 38 gastritis patients by quantitative polymerase chain reaction (qPCR). The study showed that tongue coating microbiota could potentially be a non-invasive biomarker, which might be objective and suitable for long-term monitoring.
Results
Patient and healthy control characteristics
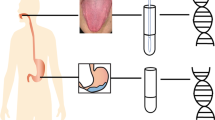
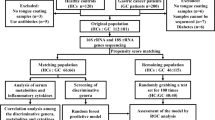
In an exploratory cohort, 78 gastritis patients received an endoscopic examination were enrolled. Gastritis patients were divided into 3 groups according to gastric precancerous cascade, including superficial gastritis, atrophic gastritis and intestinal metaplasia, based on histopathology. Fifty healthy volunteers with no complaints of stomach discomfort were recruited as normal controls (Tables 1 and S1). Tongue images and tongue-coating samples were collected from all participants. For patients, the clinical symptoms were also recorded (Table S2). In a validation cohort, 15 superficial gastritis, 7 atrophic gastritis and 16 intestinal metaplasia patients were recruited. Metagenomic sequencing was conducted in the exploratory cohort to determine the tongue-coating microbiota associated with gastritis. Campylobacter concisus was tested in tongue coating and gastric fluid in the validation cohort by qPCR (Fig. 1).
Phylogenetic and gene profiles of tongue-coating microbiomes in controls and patients
Metagenomic sequencing was performed on tongue-coating samples, generating approximately 24–211 million reads per sample. We denoted this database as Meta-Tongue database. Using a pipeline similar to the Human Microbiome Project (HMP), 176 species were obtained from all tongue coating samples. These species were classified into 12 phyla, 21 classes, 35 orders, 56 families and 87 genera (Fig. 2A and Table S3). Variations of microbial composition at the phylum level between individuals could be seen (Fig. 2B). De novo sequence assembly and gene clustering on tongue coating samples identified ~1.9 M non-redundant putative genes. The genes were annotated to 5,377 KOs and classified according to functions (Fig. 2C and Table S4).
Comparing the taxonomy of normal controls and gastritis patients, we found that 149 species were shared in normal controls and gastritis patients, and 17 species were observed only in normal controls, whereas 10 species were observed in gastritis patients. Considering the stage of patients, 8 species, 1 species and 1 species were observed only in superficial gastritis, atrophic gastritis and IM patients, respectively. Comparing the microbial constitution of HP-positive and HP-negative patients, 143 species were shared in both groups, whereas 14 species and 6 species existed only in HP-positive and HP-negative patients, respectively (Fig. S1). Considering the abundance of microbes at the phylum level, Proteobacteria, Firmicutes, Fusobacteria, Actinobacteria and Bacteroidetes dominated the tongue coating microbiota. Among which, Fusobacteria was significantly higher in patients (P = 0.003, Wilcoxon rank-sum test followed by FDR correction), whereas others had not significant differences between two groups (Fig. S2).
Tongue-coating species as a potential biomarker associated with gastritis
In terms of alpha diversity, the species richnesses in gastritis patients were significantly lower than those of normal controls (P = 0.01, Wilcoxon rank-sum test), indicating a lower number of species in gastritis patients. The Shannon indexes were significantly higher than those of normal controls (P = 0.0005, Wilcoxon rank-sum test), indicating a more uniform species distribution in gastritis patients (Fig. 3A). The alpha diversities of HP-positive and HP-negative patients did not show a significant difference (Fig. S3). In terms of beta diversity, the Jaccard distances between gastritis patients were higher than those between normal controls, indicating that patients’ samples were highly dissimilar (Fig. 3A). There was no difference of Jaccard distances between HP-positive patients and those between HP-negative patients (Fig. S4). After Wilcoxon rank-sum test followed by FDR correction, no species were found significantly different between HP-positive and HP-negative patients. After comparing patients with different spicy preferences (23 prefer spicy food, 32 do not eat spicy food, 23 no obvious preference), no species were found to be significantly different among three groups (P > 0.5, Wilcoxon rank-sum test followed by FDR correction).
Species with significantly different abundance in tongue coating of gastritis patients and normal controls. (A) Top, richness and Shannon index in normal controls and gastritis patients. Down, Jaccard distances between normal controls and between gastritis patients. (B) Top, significantly decreased abundance of species in gastritis patients. Down, significantly increased abundance of species in gastritis patients. Boxes describe the interquartile range (IQR). Red and green boxes represent the abundance of the species in normal controls and gastritis patients, respectively. The y-axis represents the log of the abundance of the corresponding species in all samples. Species from the left to the right were arranged in ascending order of P value. (C) A correlation network of species associated with gastritis. All nodes represented species that are potential biomarkers associated with gastritis. They were classified into two groups including controls-enriched species and patients-enriched species. The size of the nodes indicates the mean abundance of the species among all samples. The color of the nodes represents the taxonomy assignment, with nodes of the same color belonging to the same phylum. Edges between nodes represent the correlation between the abundance of two species. Two nodes are linked if the Spearman correlation test shows a P-value < 0.05. Solid lines represent a positive correlation, while dashed lines represent a negative correlation. The width of the edges is proportional to the correlation strength, and wide line indicates strong correlation. (D) Species whose abundance is associated with particular symptoms. Statistical comparison by Wilcoxon rank-sum test followed by false discovery rate correction. (E) A schematic diagram showing the potential functions of some species enriched in patients that have a predicted gastritis association
The Wilcoxon rank-sum test followed by FDR correction was used to characterize the significance of differences in tongue-coating microbiota abundance between the normal controls and gastritis patients. Finally, 21 species were found to be significantly different in abundance between the two groups (P < 0.05): 11 species including Veillonella parvula, Corynebacterium matruchotii, Kingella oralis, Atopobium rimae, Aggregatibacter aphrophilus, Streptococcus sanguinis, Acinetobacter lwoffii, Prevotella amnii, Prevotella bivia, Cardiobacterium hominis and Oribacterium sinus were decreased in gastritis patients, whereas 10 species including Streptococcus infantis, Treponema vincentii, Leptotrichia unclassified, Campylobacter rectus, Campylobacter showae, Capnocytophaga gingivalis, Leptotrichia buccalis, Campylobacter concisus, Selenomonas flueggei and Leptotrichia hofstadii were increased in gastritis group (Fig. 3B). These 21 species were defined as tongue-coating species associated with gastritis. To explore whether these tongue-coating species can distinguish gastritis patients from normal controls, hierarchical clustering of the abundance of these species in tongue coating samples from gastritis patients and normal controls was performed (Fig. S5). The clustering result showed that the gastritis patients group and normal controls group can be mainly separated, indicating that tongue-coating microbes may be potential biomarkers for gastritis.
A correlation network was constructed to assess the potential relationship between these 21 tongue-coating species (Fig. 3C). Controls-enriched species were significantly decreased in gastritis patients’ tongue coating, while patients-enriched species were significantly increased in gastritis patients’ tongue coating. Patients-enriched species had a stronger correlation with each other than controls-enriched species (P = 0.005, Wilcoxon rank-sum test), suggesting that patients-enriched species affected the host by interacting with each other and playing similar roles. Furthermore, the abundances of some species were also correlated with symptoms. Cardiobacterium hominis was higher in patients with bitter taste, and Selenomonas flueggei was higher in patients with dry mouth (Fig. 3D).
After exploring the potential functions of these tongue-coating species in gastritis by literature mining, we found some species enriched in patients that had the potential to induce inflammation and immune response in the host (Fig. 3E). Campylobacter concisus preferentially attached to cell-cell junctions, which led to damage of epithelial barrier functions (Man et al., 2010a, b). Campylobacter concisus could induce expression of cytokines and chemokines such as tumor necrosis factor (TNF), interleukin 1 beta (IL1B), interleukin 10 (IL10), C-C motif chemokine ligand 2 (CCL2), C-X-C motif chemokine ligand 1 (CXCL1), C-X-C motif chemokine ligand 2 (CXCL2), C-X-C motif chemokine ligand 9 (CXCL9), C-X-C motif chemokine ligand 10 (CXCL10), the assembly of inflammasome interferon gamma inducible protein 16 (IFI16) and activate the key inflammatory pathways involving nuclear factor kappa B (NF-kB), signal transducer and activator of transcription (STAT), cAMP responsive element binding protein 1 (CREB1) and interferon regulatory factor signaling (Man et al., 2010a, b; Kaakoush et al., 2015). Members of the Campylobacter concisus secretome including flagellin B (FlaB), ATP synthase F1 alpha subunit and outer membrane protein 18 (OMP18) were able to stimulate an immunoreaction (Kovach et al., 2011). Furthermore, the GroEL-like protein, which could induce the secretion of interleukin 6 (IL6) and interleukin 8 (IL8), could be secreted by Campylobacter rectus (Hinode et al., 1998). These findings showed the potential functions of species that had a predicted gastritis association.
Differential abundance of tongue-coating microbial genes between controls and patients
To explore the variation in tongue-coating microbial functions in gastritis samples, genes of tongue-coating microbes were analyzed. After comparing the genes annotated to Kyoto Encyclopedia of Genes and Genomes (KEGG) between normal controls and gastritis patients by Wilcoxon rank-sum test followed by FDR correction, we found that 878 genes were significantly different between normal controls and gastritis patients (P < 0.05). Among these, 519 genes were significantly increased in gastritis patients (Table S5), including DedD protein, threonine aldolase and N-acetylmuramoyl-l-alanine amidase. Moreover, 359 genes including adenosylcobinamide hydrolasewere, NADP-dependent alcohol dehydrogenase, putative metalloprotease and glyoxylate reductase were significantly decreased in gastritis patients (Table S6).
In order to reveal the functions that up-regulated genes played, enrichment analysis was conducted by Fisher’s exact test followed by FDR correction. Finally, 519 up-regulated genes enriched in 28 pathways (P < 0.05) (Fig. 4). Pathways such as metabolic pathways, microbial metabolism in diverse environments, biosynthesis of secondary metabolites, biosynthesis of antibiotics, flagellar assembly, bacterial chemotaxis, ABC transporters, carbon metabolism and biosynthesis of amino acids were found to be enriched in up-regulated genes, indicating a variation of microbial functions in gastritis patients.
Up-regulated tongue-coating microbial genes and enriched pathways. Blue nodes represent KEGG KOs, and yellow nodes represent KEGG pathways. The edge between two nodes indicates that the gene was in the pathway. The size of the yellow node is proportional to the connection degree, i.e., pathways with a higher number of up-regulated genes exhibit larger nodes
Campylobacter concisus is associated with gastritis stages, and can be detected in gastric fluid
To assess the differences of patients in different stages of gastric precancerous cascade, the abundances of tongue-coating microbes were compared in four groups including normal controls, superficial gastritis, atrophic gastritis and intestinal metaplasia. This division indicated the development from normal status to the transformation from gastritis to precancerous lesions. Finally, the abundance of Campylobacter concisus, together with the abundances of the class, order, family, and genus it belongs to, were found to have clear association with gastritis stages (P = 0.02, Cochran-Armitage test). The abundance of Campylobacter concisus in superficial gastritis, atrophic gastritis and intestinal metaplasia was higher than that in normal controls. In addition, the abundance of Campylobacter concisus increased during precancerous cascade (Fig. 5). The Cochran-Armitage test for trend showed that there was a significant association between the abundance of Campylobacter concisus and the precancerous cascade (P = 0.02), which was more significant when using only the first three stages (normal controls, superficial gastritis and atrophic gastritis, P = 0.004).
In the intestinal metaplasia group, the samples clearly clustered into two subclasses according to the abundance of Campylobacter concisus. We termed the subclass with high abundance as the “high abundance group” (HAG) and another subclass as the “low abundance group” (LAG). The HAG samples showed a distinct increasing trend together with the health and other two gastritis stages groups (Cochran-Armitage test for trend, P = 3.8 × 10−5). This indicated that high heterogeneity exists in the intestinal metaplasia stage and there may be subgroups with specific characteristics such as TCM or western medicine phenotypes, which requires further studies in large samples.
After analysis of the potential relationships between Campylobacter concisus and clinical symptoms, we found that gastritis patients with dry mouth had a higher abundance of Campylobacter concisus in tongue coating than others (Fig. S6).
We also tested whether Campylobacter concisus existed in the stomach. Gastric fluid samples from 38 gastritis patients with matched tongue-coating samples were collected as a validation cohort. We performed qPCR to measure the abundance of Campylobacter concisus (Table S7). Campylobacter concisus can be detected in both tongue-coating samples and gastric fluid samples.
DISCUSSION
Evidence from decades of recent long-term clinical studies and epidemiological material indicates that non-resolving inflammation has strong relationships with tumorigenesis (Carl Nathan, 2010). At least 15% of malignant tumors worldwide, especially tumors in the digestive system, respiratory system and urogenital system, are derived from non-resolving inflammation (Lisa M. Coussens, 2002). The occurrence of gastric cancer has a strong correlation with gastritis (Ohata et al., 2004). Therefore, it would be very meaningful to find biomarkers for the occurrence and development of gastritis.
Our previous study has found that gut microbiome and molecular networks are associated with the process from inflammation to cancer of colorectal cancer (Liang et al., 2014), with some inflammation-cancer related microbes validated by later studies (Kesselring et al., 2016; Zitvogel et al., 2018). In this paper, we proposed tongue-coating microbes as potential biomarkers associated with gastritis including the gastric precancerous cascade, which might be objective, non-invasive and suitable for long-term monitoring. Many researchers have revealed a correlation between oral cavity microbiota and disease in other regions of the body. James et al. compared the salivary microbiota between pancreatic cancer patients and normal controls and found that Streptococcus infantis and Campylobacter concisus were increased in pancreatic cancer patients (Farrell et al., 2012). The increase in Streptococcus infantis and Campylobacter concisus was also observed in the tongue coating of gastritis patients in our results, indicating that these potential biomarkers may also be a reflection of other disease states. A low diversity was found in tongue coating microbiota in gastritis patients, which was consistent with findings in other diseases. A low diversity in the gut was linked to obesity and inflammatory bowel disease (Turnbaugh et al., 2008; Qin et al., 2010).
Some species of the genus Campylobacter are shown to be involved in human disease, of which Campylobacter jejuni and Campylobacter coli are the most common (Man, 2011; O Brien, 2017). Campylobacter jejuni and Campylobacter coli are able to cause gastrointestinal illness including enteritis (Allos, 2001; Tam, 2003). Campylobacter concisus has been found to exist in most parts of the human digestive tract, including the oral cavity, esophagus, stomach and colon (Macfarlane et al., 2007; Zhang et al., 2010; Mukhopadhya et al., 2011), and has been repeatedly proven to be associated with gastrointestinal disease. Previous studies mostly focused on the higher isolation incidence of Campylobacter concisus in patients with inflammatory bowel disease (IBD) (Engberg et al., 2005; Newell, 2005; Man et al., 2010; Kaakoush et al., 2011; Mukhopadhya et al., 2011; Kaakoush and Mitchell, 2012; Kaakoush et al., 2014; Zhang, 2014; Deshpande et al., 2016). Campylobacter concisus is also found to be linked with Barrett’s esophagus (Macfarlane et al., 2007). Researchers focusing on microbiota in the stomach fluid showed high transcriptional active of Campylobacter concisus (von Rosenvinge et al., 2013), indicating that Campylobacter concisus may play an important role in the stomach. Our study characterized the association between the abundance of Campylobacter concisus in tongue coating and the occurrence and development of gastritis. The abundance of Campylobacter concisus in the stomach did not show significant correlation with diseases stages. There is heterogeneity in the gastritis samples, which makes it difficult to show significant correlation in small sample cohort. There might be subgroups with specific characteristics such as TCM or western medicine phenotypes, which require further studies in large samples.
There are cheaper and convenient ways for gastritis diagnosis such as endoscopy and histological examination in clinical practice, however, they are invasive. Biomarkers from tongue coating microbiome based on tongue diagnosis could provide beneficial complement for gastritis diagnosis, from the non-invasive, individualized and long-term monitoring aspects. Furthermore, in traditional Chinese medicine, tongue coating can reflect the health status of human body. Our work provided the biological evidence of tongue diagnosis. We found that species associated with gastritis had potential correlation with symptoms including dry mouth and bitter taste. In addition, Campylobacter concisus and Campylobacter rectus were recorded to upregulate genes such as immune factors, cytokines and CCL2 (Kaakoush et al., 2015), which were associated with the Hot Syndrome of gastritis in TCM (Li et al., 2007; Li et al., 2013). Especially, TNF which can be induced by Campylobacter concisus is associated with bitter taste (Feng et al., 2015), which is an important Damp Heat phenotype related to gastritis in traditional Chinese medicine. These findings showed a slight hint that tongue coating species such as Campylobacter concisus may have potential association with Hot Syndrome, which includes status such as “Shang-huo”, Damp Heat, Yin-deficiency, and so on in TCM.
In conclusion, we have demonstrated the variation of tongue-coating microbiota in gastritis patients by metagenomic sequencing, identified potential biomarkers for gastritis including precancerous cascade. Our work takes a step toward a potential non-invasive biomarker for gastritis, which might be objective and suitable for long-term monitoring. Furthermore, before using tongue-tongue microbiomes as biomarker for larger samples, more studies are required to reveal the influence of food intake and the geographical areas of people. It has been known that Helicobacter pylori is an important factor in gastritis and has been found associated with some gastritis sub-types as well as gastric cancer in many studies (McColl, 2010). Gastritis is actually an “umbrella term” for a number of diseases including those induced by Helicobacter pylori and by other factors (Sugano et al., 2015; Suzuki and Mori, 2015). Patients without Helicobacter pylori infection may also have gastritis including superficial, atrophic or even gastric cancer (Genta and Sonnenberg, 2015; Pogoriler et al., 2015; Horiuchi et al., 2016; Overby et al., 2017). In future research on larger samples, taking precise Helicobacter pylori information into consideration in the study of gastritis sub-types and stages may reveal more specific biomarkers for the diagnose of gastritis and gastric cancer.
MATERIALS AND METHODS
Sample collection
For the exploratory cohort, 99 patients who received an endoscopic examination in Beijing Dongzhimen TCM Hospital, Beijing Xiyuan TCM Hospital and China-Japan Friendship Hospital were recruited. Endoscopic examinations were performed, and the clinical symptoms were recorded. Biopsy specimens were selected from the antrum. The histological assessment was done by two experienced pathologists following clinical guidelines according to “the updated Sydney System” (Dixon et al., 1996). The inclusion criteria were a confirmed diagnosis of gastritis according to histological examination. Autoimmune gastritis patients were excluded. The Helicobacter pylori infection for all patients was identified by pathological examination. Healthy volunteers were recruited from Tsinghua University during their annual physical examination in the hospital. Seventy-six healthy people who reported no complaints of stomach discomfort in the past 5 years and also confirmed no gastritis in the examination were enrolled. The exclusion criteria for both patients and healthy volunteers consisted of the use of glucocorticoids and antibiotics for the past 3 months. Tongue images and tongue coating samples were collected from all participants. After quality control of metagenomic sequencing, 167 samples including 93 patients and 74 controls passed the acceptance criteria. Samples with contamination and patients without histopathological results were then filtered. Finally, 78 patients and 50 controls were retained for further analysis. Patients were divided into different stages of gastric precancerous cascade according to the histopathological results. This study mainly focused on people living in Beijing.
For the validation cohort, 38 patients who received an endoscopic examination in the China-Japan Friendship Hospital were recruited with the same inclusion and exclusion criteria for the exploration cohort. Samples were different from that in exploratory cohort. Tongue-coating and gastric fluid samples were collected. Histological examination results and clinical symptoms were recorded.
Specimen collection, DNA extraction and preservation
Tongue-coating swabs were used to collect tongue-coating samples of participants before consumption of breakfast and water. The tongue was scraped from the root to the tip 30 times by simultaneously rolling the swab, the swab was placed into an RNase-free Eppendorf tube with 1 mL of phosphate-buffered saline (PBS), and the swab was agitated in order to wash out the tongue coating. The above step was repeated twice with a new swab and new Eppendorf tube to ensure that the tongue coating was sufficiently collected. After collection of tongue coating, the tubes were centrifuged at 5,000 ×g for 5 min. Supernatant and sediment were preserved separately in sterile tubes and stored at −80 °C until analysis. DNA of microbiota in the samples was extracted following the user guide of a modified version of the MO-BIO PowerSoil DNA Isolation Kit (MO-BIO Laboratories, Inc., Carlsbad, CA, USA).
Library construction and sequencing
The sequencing library was constructed using a modified version of the NEBNext Ultra DNA library Prep Kit (New England Biolabs, Ipswich, MA, USA) protocol. We constructed barcoded, paired-end libraries with an insert size of ~500 bp for each sample. Four samples were designed to be sequenced in each lane. Samples were randomly assigned to different sequence lanes. Extracted DNA (200 ng) was used for library construction for each sample. Samples were sequenced in two batches. Samples in the first batch (172 samples) were sent to BGI Shenzhen for sequencing. After quality determination, libraries passing quality control were sequenced with the Illumina HiSeq 2000 platform. The read length was set to 90 bp. Samples in the second batch (3 samples) were sequenced in Tsinghua University with the Illumina HiSeq 2500 platform. The read length was set to 100 bp.
Quality control, assembly and gene prediction
A modified version of fastx_barcode_splitter in the FASTX-Toolkit (http://hannonlab.cshl.edu/fastx_toolkit/) was used to decode the barcoded samples of each lane with the parameter “—mismatches 2”. We used the pipeline “assemble_revise_predict_genes_with_hg19_screen” in the “runMOCAT.pl” of the MOCAT V1.3 (http://vm-lux.embl.de/~kultima/MOCAT/) (Kultima et al., 2012) toolkit for quality control, assembly and gene (ORF) prediction. The configured file of MOCAT was similar to that used by Li et al. (2014).
Sequences after quality control were clean reads of high quality without human reads. To ensure better reliability, only samples with more than 15 M clean reads were used for subsequent analysis. A total of 167 samples passed the entire quality control (Table S8).
Taxonomy profiling
We used MetaPhlAn Version 1.7.8 (http://huttenhower.sph.harvard.edu/metaphlan) (Segata et al., 2012) to calculate the taxonomy abundance table for each sample. MetaPhlAn aligns short reads directly to the customized prokaryote database, which was constructed by extracting clade-specific marker genes from the NCBI genome database with the phylogenetic information of the NCBI taxonomic tree. Abundances were calculated based on read counts of corresponding marker genes normalized by gene length and sequencing depth. We used clean reads as inputs, and the parameters were “–bt2_ps very-sensitive, -t ‘rel_ab’”.
Construction and annotation of Integrated Tongue Gene Catalogue (ITGC)
We first constructed a “Meta-Tongue Gene Catalogue” (MTGC) for the Meta-Tongue dataset. We merged all the ORFs from 167 high quality samples and did a redundancy removal according to their sequence similarity using CD-HIT (Li and Godzik, 2006; Fu et al., 2012) v4.6.1 (http://weizhongli-lab.org/cd-hit/). The parameters we used in CD-HIT were “-G 0 -M 0 -T 4 -c 0.95 -aS 0.9 -n 8 -B 1”. To construct a more comprehensive catalogue, we collected HMP dorsum samples (Huttenhower et al., 2012) and used a same pipeline as above to build an “HMP Tongue Gene Catalogue” (HTGC). We then merged MTGC and HTGC using CD-HIT with same parameters, and got the final Integrated Tongue Gene Catalogue (ITGC).
We mapped ITGC genes to NCBI nr database (created in 2012.2.28 by NCBI) with blastp (Altschul et al., 1990) in the Blast 2.2.29+ toolkit. We used “-evalue 1e-5 -outfmt 6” in our study. The outputs from blastp were processed with MEGAN (Huson et al., 2007) v5.7.1 (http://ab.inf.uni-tuebingen.de/software/megan5/) with parameters “maxMatches=100 minScore=50.0 maxExpected=0.01 topPercent=10.0 minSupport=1 minComplexity=0.0 useMinimalCoverageHeuristic=false paired=false useIdentityFilter=false”.
Gene and function profiling
For each sample, we mapped its clean paired-end reads to ITGC genes, calculated gene abundances and function abundances according to the mapping results. We used SOAPaligner v2.2.1 (http://soap.genomics.org.cn/soapaligner.html) with parameters “-r 2 -M 4 -l 30 -v 9” in our study. The ITGC gene relative abundances were calculated in a similar way with that in the work of Qin et al. (2012).
Statistical analysis
To determine the differential abundance of species and genes between controls and patients, the Wilcoxon rank-sum test was used. P-values were adjusted with false discovery rate. The correlation between species was analyzed using Spearman’s correlation. The comparison of alpha diversity index and Jaccard distance was tested by Wilcoxon rank-sum test. The increasing trend of the abundance of Campylobacter concisus with gastritis stages was tested by Cochran-Armitage test. Samples in HAG were labeled as 1 and others were labeled as 0. Health and the three gastritis stages were labeled as 0, 1, 2, 3 separately.
Detection of Campylobacter concisus by qPCR
Abundances of Campylobacter concisus in tongue coating and gastric fluid were assessed using quantitative PCR (qPCR). Universal 16S rDNA was used as internal reference for the estimation of all the microbes. The abundance of Campylobacter concisus was estimated using the proportion of Campylobacter concisus in all the microbes. qPCR was performed with TransStart Top Green Qpcr SuperMix.
References
Allos BM (2001) Campylobacter jejuni Infections: update on emerging issues and trends. Clin Infect Dis 32:1201–1206
Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ (1990) Basic local alignment search tool. J Mol Biol 215:403–410
Aviles-Jimenez F, Vazquez-Jimenez F, Medrano-Guzman R, Mantilla A, Torres J (2014) Stomach microbiota composition varies between patients with non-atrophic gastritis and patients with intestinal type of gastric cancer. Sci Rep 4:4202
Carl Nathan AD (2010) Nonresolving inflammation. Cell. 19:871–882
Coker OO, Dai Z, Nie Y, Zhao G, Cao L, Nakatsu G, Wu WK, Wong SH, Chen Z, Sung JJY et al (2017) Mucosal microbiome dysbiosis in gastric carcinogenesis. Gut:2017–314281
Correa P (1992) Human gastric carcinogenesis: a multistep and multifactorial process—1st American-cancer-society award lecture on cancer-epidemiology and prevention. Cancer Res 52:6735–6740
Correa P, Piazuelo MB (2012) The gastric precancerous cascade. J Digest Dis 13:2–9
Coussens LM, Werb Z (2002) Inflammation and cancer. Nature. 420:860
Deshpande NP, Wilkins MR, Castaño-Rodríguez N, Bainbridge E, Sodhi N, Riordan SM, Mitchell HM, Kaakoush NO (2016) Campylobacter concisus pathotypes induce distinct global responses in intestinal epithelial cells. Sci Rep 6:34288
Dixon MF, Genta RM, Yardley JH, Correa P (1996) Classification and grading of gastritis: The updated sydney system. Am J Surg Pathol 20:1161–1181
Engberg J, Bang DD, Aabenhus R, Aarestrup FM, Fussing V, Gerner-Smidt P (2005) Campylobacter concisus: an evaluation of certain phenotypic and genotypic characteristics. Clin Microbiol Infect 11:288–295
Eun CS, Kim BK, Han DS, Kim SY, Kim KM, Choi BY, Song KS, Kim YS, Kim JF (2014) Differences in gastric mucosal microbiota profiling in patients with chronic gastritis, intestinal metaplasia, and gastric cancer using pyrosequencing methods. Helicobacter 19:407–416
Farrell JJ, Zhang L, Zhou H, Chia D, Elashoff D, Akin D, Paster BJ, Joshipura K, Wong DTW (2012) Variations of oral microbiota are associated with pancreatic diseases including pancreatic cancer. Gut 61:582–588
Feng P, Jyotaki M, Kim A, Chai J, Simon N, Zhou M, Bachmanov AA, Huang L, Wang H (2015) Regulation of bitter taste responses by tumor necrosis factor. Brain Behav Immunol 49:32–42
Ferreira RM, Pereira-Marques J, Pinto-Ribeiro I, Costa JL, Carneiro F, Machado JC, Figueiredo C (2017). Gastric microbial community profiling reveals a dysbiotic cancer-associated microbiota. Gut:2017–314205
Filipe MI, Munoz N, Matko I, Kato I, Pompe-Kirn V, Jutersek A, Teuchmann S, Benz M, Prijon T (1994) Intestinal metaplasia types and the risk of gastric cancer: a cohort study in Slovenia. Int J Cancer 57:324–329
Fu L, Niu B, Zhu Z, Wu S, Li W (2012) CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics 28:3150–3152
Genta RM, Sonnenberg A (2015) Helicobacter-negative gastritis: a distinct entity unrelated to Helicobacter pylori infection. Aliment Pharmacol Ther 41:218–226
Guo Y, Nie Q, MacLean AL, Li Y, Lei J, Li S (2017) Multiscale modeling of inflammation-induced tumorigenesis reveals competing oncogenic and oncoprotective roles for inflammation. Cancer Res 77:6429–6441
Hakalehto E, Vilpponen-Salmela T, Kinnunen K, von Wright A (2011) Lactic acid bacteria enriched from human gastric biopsies. ISRN Gastroenterol 2011:1–4
Hinode D, Yoshioka M, Tanabe S, Miki O, Masuda K, Nakamura R (1998) The GroEL-like protein from Campylobacter rectus: immunological characterization and interleukin-6 and -8 induction in human gingival fibroblast. FEMS Microbiol Lett 167:1–6
Horiuchi Y, Fujisaki J, Yamamoto N, Shimizu T, Miyamoto Y, Tomida H, Taniguchi C, Morishige K, Omae M, Ishiyama A et al (2016) Biological behavior of the intramucosal Helicobacter pylori-negative undifferentiated-type early gastric cancer: comparison with Helicobacter pylori-positive early gastric cancer. Gastric Cancer 19:160–165
Huson DH, Auch AF, Qi J, Schuster SC (2007) MEGAN analysis of metagenomic data. Genome Res 17:377–386
Huttenhower C, Gevers D, Knight R, Abubucker S, Badger JH, Chinwalla AT, Creasy HH, Earl AM, FitzGerald MG, Fulton RS et al (2012) Structure, function and diversity of the healthy human microbiome. Nature 486:207–214
Jiang B, Liang X, Chen Y, Ma T, Liu L, Li J, Jiang R, Chen T, Zhang X, Li S (2012) Integrating next-generation sequencing and traditional tongue diagnosis to determine tongue coating microbiome. Sci Rep 2:936
Jonkers D, Gisbertz I, de Bruine A, Bot F, Arends JW, Stobberingh E, Schouten H, Stockbrugger R (1997) Helicobacter pylori and non-Helicobacter pylori bacterial flora in gastric mucosal and tumour specimens of patients with primary gastric lymphoma. Eur J Clin Invest 27:885–892
Kaakoush NO, Mitchell HM (2012) Campylobacter concisus: a new player in intestinal disease. Front Cell Infect MI:2
Kaakoush NO, Deshpande NP, Wilkins MR, Tan CG, Burgos-Portugal JA, Raftery MJ, Day AS, Lemberg DA, Mitchell H (2011) The pathogenic potential of Campylobacter concisus strains associated with chronic intestinal diseases. PLoS ONE 6:e29045
Kaakoush NO, Mitchell HM, Man SM (2014) Role of emerging campylobacter species in inflammatory bowel diseases. Inflamm Bowel Dis 20:2189–2197
Kaakoush NO, Deshpande NP, Man SM, Burgos-Portugal JA, Khattak FA, Raftery MJ, Wilkins MR, Mitchell HM (2015) Transcriptomic and proteomic analyses reveal key innate immune signatures in the host response to the gastrointestinal pathogen Campylobacter concisus. Infect Immunol 83:832–845
Kanawong R, Obafemi-Ajayi T, Ma T, Xu D, Li S, Duan Y (2012) Automated tongue feature extraction for ZHENG classification in traditional chinese medicine. Evid-Based Complement Altern 2012:1–14
Kesselring R, Glaesner J, Hiergeist A, Naschberger E, Neumann H, Brunner SM, Wege AK, Seebauer C, Köhl G, Merkl S et al (2016) IRAK-M expression in tumor cells supports colorectal cancer progression through reduction of antimicrobial defense and stabilization of STAT3. Cancer Cell 29:684–696
Kovach Z, Kaakoush NO, Lamb S, Zhang L, Raftery MJ, Mitchell H (2011) Immunoreactive proteins of Campylobacter concisus, an emergent intestinal pathogen. FEMS Immunol Med Microbiol 63:387–396
Kultima JR, Sunagawa S, Li J, Chen W, Chen H, Mende DR, Arumugam M, Pan Q, Liu B, Qin J et al (2012) MOCAT: a metagenomics assembly and gene prediction toolkit. PLoS ONE 7:e47656
Li W, Godzik A (2006) Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 22:1658–1659
Li S, Zhang ZQ, Wu LJ, Zhang XG, Li YD, Wang YY (2007) Understanding ZHENG in traditional Chinese medicine in the context of neuro-endocrine-immune network. IET Syst Biol 1:51–60
Li X, Wong GL, To K, Wong VW, Lai LH, Chow DK, Lau JY, Sung JJ, Ding C (2009) Bacterial microbiota profiling in gastritis without Helicobacter pylori infection or non-steroidal anti-inflammatory drug use. PLoS ONE 4:e7985
Li R, Ma T, Gu J, Liang X, Li S (2013) Imbalanced network biomarkers for traditional Chinese medicine Syndrome in gastritis patients. Sci Rep 3:1543
Li J, Jia H, Cai X, Zhong H, Feng Q, Sunagawa S, Arumugam M, Kultima JR, Prifti E, Nielsen T et al (2014) An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol 32:834–841
Liang X, Li H, Tian G, Li S (2014) Dynamic microbe and molecule networks in a mouse model of colitis-associated colorectal cancer. Sci Rep 4:4985
Macfarlane S, Furrie E, Macfarlane GT, Dillon JF (2007) Microbial colonization of the upper gastrointestinal tract in patients with Barrett’s esophagus. Clin Infect Dis 45:29–38
Man SM (2011) The clinical importance of emerging Campylobacter species. Nat Rev Gastroenterol Hepatol 8:669–685
Man SM, Kaakoush NO, Leach ST, Nahidi L, Lu HK, Norman J, Day AS, Zhang L, Mitchell HM (2010a) Host attachment, invasion, and stimulation of proinflammatory cytokines by Campylobacter concisus and other non-Campylobacter jejuni Campylobacter Species. J Infect Dis 202:1855–1865
Man SM, Zhang L, Day AS, Leach ST, Lemberg DA, Mitchell H (2010b) Campylobacter concisus and other Campylobacter species in children with newly diagnosed Crohnʼs disease. Inflamm Bowel Dis 16:1008–1016
Marshall BJ, Warren JR (1984) Unidentified curved bacilli in the stomach of patients with gastritis and peptic ulceration. Lancet 1:1311–1315
McColl KE (2010) Clinical practice. Helicobacter pylori infection. Eur J Pediatr 362:1597–1604
Meining A, Riedl B, Stolte M (2002) Features of gastritis predisposing to gastric adenoma and early gastric cancer. J Clin Pathol 55:770–773
Miehlke S, Hackelsberger A, Meining A, Hatz R, Lehn N, Malfertheiner P, Stolte M, Bayerdorffer E (1998) Severe expression of corpus gastritis is characteristic in gastric cancer patients infected with Helicobacter pylori. Br J Cancer 78:263–266
Mukhopadhya I, Thomson JM, Hansen R, Berry SH, El-Omar EM, Hold GL (2011) Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS ONE 6:e24190
Newell DG (2005) Campylobacter concisus: an emerging pathogen? Eur J Gastroenterol Hepatol 17:1013–1014
O Brien SJ (2017) The consequences of Campylobacter infection. Curr Opin Gastroen 33:14–20
Ohata H, Kitauchi S, Yoshimura N, Mugitani K, Iwane M, Nakamura H, Yoshikawa A, Yanaoka K, Arii K, Tamai H et al (2004) Progression of chronic atrophic gastritis associated with Helicobacter pylori infection increases risk of gastric cancer. Int J Cancer 109:138–143
Overby A, Murayama SY, Michimae H, Suzuki H, Suzuki M, Serizawa H, Tamura R, Nakamura S, Takahashi S, Nakamura M (2017) Prevalence of gastric non-Helicobacter pylori-Helicobacters in Japanese patients with gastric disease. Digestion 95:61–66
Owen DA (2003) Gastritis and carditis. Mod Pathol 16:325–341
Pogoriler J, Kamin D, Goldsmith JD (2015) Pediatric non-Helicobacter pylori atrophic gastritis: a case series. Am J Surg Pathol 39:786–792
Price AB (1991) The Sydney system: histological division. J Gastroenterol Hepatol 6:209–222
Qin J, Li R, Raes J, Arumugam M, Burgdorf KS, Manichanh C, Nielsen T, Pons N, Levenez F, Yamada T et al (2010) A human gut microbial gene catalogue established by metagenomic sequencing. Nature 464:59–65
Qin J, Li Y, Cai Z, Li S, Zhu J, Zhang F, Liang S, Zhang W, Guan Y, Shen D et al (2012) A metagenome-wide association study of gut microbiota in type 2 diabetes. Nature 490:55–60
Rugge M, Meggio A, Pennelli G, Piscioli F, Giacomelli L, De Pretis G, Graham DY (2007) Gastritis staging in clinical practice: the OLGA staging system. Gut 56:631–636
Sahay P, West AP, Birkenhead D, Hawkey PM (1995) Campylobacter jejuni in the stomach. J Med Microbiol 43:75–77
Schulz C, Schütte K, Malfertheiner P (2016) Helicobacter pylori and other gastric microbiota in gastroduodenal pathologies. Digest Dis 34:210–216
Segata N, Waldron L, Ballarini A, Narasimhan V, Jousson O, Huttenhower C (2012) Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods 9:811–814
Sipponen P, Kekki M, Haapakoski J, Ihamaki T, Siurala M (1985) Gastric cancer risk in chronic atrophic gastritis: statistical calculations of cross-sectional data. Int J Cancer 35:173–177
Sjostedt S, Heimdahl A, Kager L, Nord CE (1985) Microbial colonization of the oropharynx, esophagus and stomach in patients with gastric diseases. Eur J Clin Microbiol 4:49–51
Sohn SH, Kim N, Jo HJ, Kim J, Park JH, Nam RH, Seok YJ, Kim YR, Lee DH (2017) Analysis of gastric body microbiota by pyrosequencing: possible role of bacteria other than helicobacter pylori in the gastric carcinogenesis. J Cancer Prev 22:115–125
Song H, Ekheden IG, Zheng Z, Ericsson J, Nyren O, Ye W (2015) Incidence of gastric cancer among patients with gastric precancerous lesions: observational cohort study in a low risk Western population. BMJ-Brit Med J 351:3867
Stolte M, Meining A (2001) The updated Sydney system: classification and grading of gastritis as the basis of diagnosis and treatment. Can J Gastroenterol 15:591–598
Sugano K, Tack J, Kuipers EJ, Graham DY, El-Omar EM, Miura S, Haruma K, Asaka M, Uemura N, Malfertheiner P (2015) Kyoto global consensus report on Helicobacter pylori gastritis. Gut 64:1353–1367
Sun ZM, Zhao J, Qian P, Wang YQ, Zhang WF, Guo CR, Pang XY, Wang SC, Li FF, Li Q (2013) Metabolic markers and microecological characteristics of tongue coating in patients with chronic gastritis. BMC Complement Altern Med 13:227
Suzuki H, Mori H (2015) Helicobacter pylori: Helicobacter pylori gastritis: a novel distinct disease entity. Nat Rev Gastroenterol Hepatol 12:556–557
Tam C (2003) Campylobacter coli: an important foodborne pathogen. J Infect 47:28–32
Turnbaugh PJ, Hamady M, Yatsunenko T, Cantarel BL, Duncan A, Ley RE, Sogin ML, Jones WJ, Roe BA, Affourtit JP et al (2008) A core gut microbiome in obese and lean twins. Nature 457:480–484
von Rosenvinge EC, Song Y, White JR, Maddox C, Blanchard T, Fricke WF (2013) Immune status, antibiotic medication and pH are associated with changes in the stomach fluid microbiota. ISME J 7:1354–1366
Ye J, Cai X, Yang J, Sun X, Hu C, Xia J, Shen J, Su K, Yan H, Xu Y et al (2016) Bacillus as a potential diagnostic marker for yellow tongue coating. SCI REP-UK:6
Zhang L (2014) Campylobacter concisus and inflammatory bowel disease. World J Gastroenterol 20:1259
Zhang L, Budiman V, Day AS, Mitchell H, Lemberg DA, Riordan SM, Grimm M, Leach ST, Ismail Y (2010) Isolation and detection of Campylobacter concisus from saliva of healthy individuals and patients with inflammatory bowel disease. J Clin Microbiol 48:2965–2967
Zitvogel L, Ma Y, Raoult D, Kroemer G, Gajewski TF (2018) The microbiome in cancer immunotherapy: Diagnostic tools and therapeutic strategies. Science 359:1366–1370
Ethics approval and consent to participation
This study was approved by the Medical Ethical Committee of the Beijing Dongzhimen TCM Hospital and complied with the standards indicated by the Declaration of Helsinki. All participants signed the informed consent.
Availability of data and material
The Meta-Tongue metagenomics sequencing data have been deposited in BIGD database (http://bigd.big.ac.cn) and can be found under accession PRJCA000663. Gene and function abundance tables can be found in ftp://166.111.5.235:8082/Publications/Meta-Tongue.
Acknowledgements
We thank Yongan Ye and Zhihong Li in Beijing Dongzhimen TCM Hospital and Zhenhua Li in Beijing Xiyuan TCM Hospital for providing help with sample collection. We also acknowledge Lai Wei in Sun Yat-sen University and Jingren Zhang and Juanjuan Wang in Tsinghua University for their helpful advices on experiments, and thank Geng Tian and Jidong Lang in Tsinghua University and Ting Chen, Rui Jiang, Congmin Zhu, Peng Zhang and Lianshuo Li in our Division for their suggestions. We also thank Jianhuo Fang in Tsinghua University and Huiying Li, Xujun Liang, Aidi Tan and Liang Ruan in our Division for helping for sample collection. This study was supported by the National Natural Science Foundation of China (Grant Nos. 81630103, 91729301, 91229201 and 81225025) and the Project of Tsinghua-Fuzhou Insititute for Data Technology (TFIDT2018001) to S. Li and grants 61673231 and 61721003 to X. Zhang.
Abbreviations
AG, atrophic gastritis; CCL2, C-C motif chemokine ligand 2; CREB1, cAMP responsive element binding protein 1; CXCL1, C-X-C motif chemokine ligand 1; CXCL2, C-X-C motif chemokine ligand 2; CXCL9, C-X-C motif chemokine ligand 9; CXCL10, C-X-C motif chemokine ligand 10; FlaB, flagellin B; GC, gastric cancer; HAG, high abundance group; HMP, Human Microbiome Project; HP, helicobacter pylori; HTGC, HMP Tongue Gene Catalogue; IBD, inflammatory bowel disease; IFI16, interferon gamma inducible protein 16; IL1B, interleukin 1 beta; IL6, interleukin 6; IL8, interleukin 8; IL10, interleukin 10; IM, intestinal metaplasia; IQR, interquartile range; ITGC, Integrated Tongue Gene Catalogue; KEGG, Kyoto Encyclopedia of Genes and Genomes; LAG, low abundance group; MTGC, Meta-Tongue Gene Catalogue; NF-kB, nuclear factor kappa B; OMP18, outer membrane protein 18; PBS, phosphate-buffered saline; qPCR, quantitative polymerase chain reaction; SG, superficial gastritis; STAT, signal transducer and activator of transcription; TCM, traditional Chinese medicine; TNF, tumor necrosis factor
Compliance with Ethics Guidelines
Jiaxing Cui, Hongfei Cui, Mingran Yang, Shiyu Du, Junfeng Li, Yingxue Li, Liyang Liu, Xuegong Zhang and Shao Li declare that they have no conflict of interest. All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000 (5). Informed consent was obtained from all patients for being included in the study.
Author Contributions
J. Cui and H. Cui performed the bioinformatics analyses, and prepared figures and tables. M. Yang performed the qPCR experiment. J. Cui, M. Yang, L. Liu and S. Du collected the samples. H. Cui, J. Li and Y. Li processed the sequence data. S. Li and X. Zhang designed the project. S. Li conceived the study. J. Cui wrote early drafts, and all authors contributed to the revision of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Cui, J., Cui, H., Yang, M. et al. Tongue coating microbiome as a potential biomarker for gastritis including precancerous cascade. Protein Cell 10, 496–509 (2019). https://doi.org/10.1007/s13238-018-0596-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13238-018-0596-6