Abstract
Cells sense various in vivo mechanical stimuli, which initiate downstream signaling to mechanical forces. While a body of evidences is presented on the impact of limited mechanical regulators in past decades, the mechanisms how biomechanical responses globally affect cell function need to be addressed. Complexity and diversity of in vivo mechanical clues present distinct patterns of shear flow, tensile stretch, or mechanical compression with various parametric combination of its magnitude, duration, or frequency. Thus, it is required to understand, from the viewpoint of mechanobiology, what mechanical features of cells are, why mechanical properties are different among distinct cell types, and how forces are transduced to downstream biochemical signals. Meanwhile, those in vitro isolated mechanical stimuli are usually coupled together in vivo, suggesting that the different factors that are in effect individually could be canceled out or orchestrated with each other. Evidently, omics analysis, a powerful tool in the field of system biology, is advantageous to combine with mechanobiology and then to map the full-set of mechanically sensitive proteins and transcripts encoded by its genome. This new emerging field, namely mechanomics, makes it possible to elucidate the global responses under systematically-varied mechanical stimuli. This review discusses the current advances in the related fields of mechanomics and elaborates how cells sense external forces and activate the biological responses.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Mechanical stimuli are crucial to many biological processes at organ, tissue, cell, and molecule levels. Shear flow, tensile stretch, and mechanical compression are most typical in vivo mechanical stimuli, which are in action alone or synergistically with other mechanical and even biochemical factors (Wang, 2006; Cohen and Chen, 2008). For example, endothelial cells are subjected to blood shear flow and orientated towards the flow direction (Silkworth and Stehbens, 1975), extracellular matrices (ECMs) are stretched to mediate the outside-in signaling (Wright et al., 1997), and cartilage tissue is compressed to initiate interstitial fluid pressurization (Soltz and Ateshian, 2000). A body of cues is known about how these mechanical stimuli affect cell morphology, proliferation, and differentiation. Evidently, it is difficult to elucidate what really happen at cellular and molecular levels if only a single type of mechanical stimuli or one set of mechanical parameters at a given stimulus is used.
Homeostatic imbalances are main driving forces to initiate physiological changes, in which various stimuli are monitored closely by receptors and sensors at different sites. Most of mechanoreceptors often react to shear flow, tensile stretch, or mechanical compression. When multiple mechanical stimuli or parameters are pooled together, part(s) of those molecular events presented in individual stimuli tests might be reserved constantly, fostered cooperatively, or canceled out each other, since the cross-talks existing in the mechanically sensitive genes and proteins would exert null, synergistic, or opposite effects. Thus, global mapping of activated genes and proteins that are responsible to the specific mechanical stimuli is required to conduct from the viewpoint of omics. In this review, cellular and molecular responses to mechanical stimuli were discussed, and the new conceptual terminology of mechanomics referred to transcriptomics and proteomics combined with systematically-varied mechanical stimuli was proposed.
Mechanobiology and Transcriptomics/Proteomics
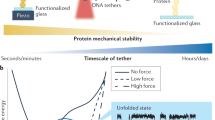
Mechanobiology is an interdisciplinary field at the interface of mechanics and biology that emerges over last two decades (Wang et al., 2008). It focuses on elucidating the mechanisms how external forces or changes in cell or tissue mechanical environment contribute to development, physiology, and disease of an organism. A major challenge in this field is to elucidate the molecular mechanisms of mechanotransduction, by which cells sense and respond to biomechanical signals and convert them into biochemical signals (Katsumi et al., 2004; Long et al., 2011; Dado et al., 2012). Numerous mechanoreceptors, such as ECM molecules, transmembrane proteins, cytoskeleton, nuclei, and lipid bilayer, are mechanically sensitive and then initiate inside-out or outside-in mechanotransduction for cells (Janmey and McCulloch, 2007). A single type of mechanical stimuli in vitro often provokes cell’s sensing and responding to external forces via multiple molecular events (Fig. 1A). By contrast, multiple types of mechanical stimuli are present around a cell or cells in vivo and act on the cell(s) synergistically (Fig. 1B). It is also noticed that the physiological stimuli are usually dynamic rather than static, which highly depends on the magnitude, frequency, and duration of mechanical forces.
Schematic of cellular responses to mechanical stimuli in one-to-more (A) or more-to-one pattern (B). Each row of the heatmap represents different genes, of which the abundance is indicated by top-right color key, and each column of the heatmap denotes distinct cells (A) and different mechanical forces (B)
On the other hand, omics is the most striking tool to globally map the cellular and molecular responses to various biochemical and biomechanical stimuli. It aims, from the viewpoint of system biology, at the characterization and quantification of pooled biological molecules that transmit external signals to alter the structure, function, and dynamics of an organism or organisms. Integration of existing approaches in biomechanics and high-throughput transcriptomics/proteomics enables us to profile cell phenotype under complicated mechanical stimuli and to elucidate the mechanisms in cell type-specific mechanobiology (Davies et al., 2005). Current efforts mainly focus on understanding the transcriptomics/proteomics on the pattern or parametric dependence of one specific type or on the combined types in respective pattern or parameter sets. Here we discuss the combination of these two fields along typical mechanical clues where the detailed parameters are summarized in Table 1 for clarity.
Mechanical modeling of a cell
Cells sense their mechanical environment, which, in turn, regulates their functions. Various theoretical models are proposed to understand mechanical regulation of cell functions, which provide differential mathematical solutions. At subcellular level, most models are attempting to predict the force or stress distribution of cytoskeletal network and mimic the remodeling of mechanically sensitive cytoskeletal proteins (Ingber, 1997; Li, 2008; Geiger et al., 2009). At cellular level, a cell is usually modeled as an encapsulated lipid membrane containing stress-supported structures to support its deformation, adhesion, and spreading (Evans and Yeung, 1989; Stamenovic and Ingber, 2002; Murrell et al., 2011). At tissue level, mathematically models are developed to predict the dynamics of tumor growth (Chaplain et al., 2006) and osteogenic differentiation (Carter et al., 1998) under mechanical stimuli. These models also help to elucidate the mechanisms how cells resist shape distortion and maintain their structural stability and how they convert mechanical signals into biochemical responses. Regardless of their advantages in theoretical prediction, these models are still required to compare with the relevant experimental measurements.
Single type of mechanical stimuli
Current works on mechanobiology and mechanotransduction are mainly focused on understanding how the cells sense and respond to a single type of mechanical stimuli and what the functional molecules are only on the basis of a few protein effectors.
Shear flow
Cellular responses to shear flow that mimics the physiological blood or interstitial flow are extensively investigated (Liang et al., 2008; Cui et al., 2011; Fu et al., 2011a; Kang et al., 2012). Exposure of live cells to shear flow induces remarkable changes in cell morphology, adhesion, and spreading (Yang et al., 2009; Zhan et al., 2012). For example, endothelial cells (ECs) display adaptive remodeling in response to shear stress, under which surface mechanosensors of integrins, Flk-1, ion channels, GPCRs, and PECAM-1 are involved in sensing shear stress (Li et al., 2005; Barakat and Lieu, 2003). Steady flow enforces myeloma cells to form actin-rich but microtubule-lacking protrusions in a stress-dependent pattern (Porat et al., 2011). Osteocytes subjected to pulsating flow appear to be more responsive than osteoblasts or periosteal fibroblast osteocytes in inhibiting osteoclast formation and resorption via NO-dependent pathways (Tan et al., 2007). Shear-induced transportation of circulating tumor cells to the target site is the prerequisite for organ-specific metastasis under blood flow (Wirtz et al., 2011) and tumor cell invasion is usually promoted by interstitial fluid affecting their interactions with stromal cells (Shieh et al., 2011).
Transcriptomic analysis is growing up in cell mechanobiology studies under fluid flow in the past decades. A specific pattern or parameter setting of shear flow initiates the expression of multiple genes in various biological processes. For instance, gene expression profile of ECs subjected to laminar or steady flow proposes the significant modulation of the genes involved in cell proliferation, ECM/cytoskeleton remodeling, angiogenesis, or in inflammatory cytokines, stress response proteins, and signaling molecules (Chen et al., 2001). Oscillatory flow up-regulates the transcription factor expression (Runx2, Sox9, and PPARγ) and induces osteogenic differentiation via RhoA and its effector protein ROCK II for the fate determination of mesenchymal stem cells (MSCs) (Arnsdorf et al., 2009). Meanwhile, different patterns or parameter settings of shear flow promote the distinct transcriptomic alternations since distinct gene profiles and resultant phenotypes of ECs are observed between low-shear disturbed flow and high-shear laminar flow to identify >100 new genes (Brooks et al., 2002). Moreover, the physiological conditions or biochemical microenvironments that cells reside should be taken into account under blood flow. One example is that the monoculture of ECs up-regulates ICAM-1 expression but the co-culture of ECs and SMCs down-regulates ICAM-1 expression under laminar flow (Heydarkhan-Hagvall et al., 2006). TNF-α-induced gene expression is distinct from that induced by disturbed flow, implying that the downstream effect of disturbed flow is not mediated as same as the signaling pathways that activate NF-κB (Brooks et al., 2002).
While much progresses have been achieved in transcriptomic analysis under shear flow, a few works are also done in proteomic profiling of cells under shear stress. Under steady flow, 10 and 3 proteins are found to be up- and down-regulated for hMSCs, respectively, in which annexin A2 and GAPDH are substantially increased (Yi et al., 2010). Besides, cells can also sense shear stress and convert it into secretary signals, as seen in 43 differential proteins found from low shear-induced rat thoracic aorta in which two secretary molecules of PDGF-BB and TGF-β1 are critical in vascular remodeling (Qi et al., 2011). Again, the combination of biochemical factors with biomechanical stress is critical, since high glucose alone significantly modulates shear-induced mechanosensing complexes and protein phosphorylation pathways of endothelium while both laminar shear and high glucose together enhance HSPs and protein ubiquitination of bovine ECs (Wang et al., 2009).
Tensile stretch
Mechanical stretch usually induces significant changes in cellular responses and tissue remodeling at molecular and cellular levels (Tamura et al., 2001; Zhou et al., 2005; Richard et al., 2007). Static stretch induces asymmetric migration of keratinocytes cocultured with fibroblasts in a wound repairing model (Lü et al., 2013). Tendon fibroblasts sense cyclic stretch in the dose- and time-dependent patterns and induce protein productions (Col-I, TGF-β1, COX, PGE2, and LTB4) (Wang, 2006). Equi-biaxial stretch promotes higher pro-MMP-2 production and its active form in anterior cruciate ligament than those in medial collateral ligament fibroblasts but no difference in post-translational modification is observed in between (Zhou et al., 2005). Transient increase of cysteine-rich protein 61 (Cyr61) mRNA is observed for fetal bovine bladder smooth muscle cells (SMCs) subjected to cyclic biaxial stretch, which is correlated with intracellular signaling (PKC, PI3K, and Rho kinase) (Tamura et al., 2001). Moreover, ion channels could serve as candidate mechanosensors, since cyclic stretch is able to trigger the gadolinium-sensitive stretch-activated ion channels, inducing a rapid Ca2+ influx, and then play a crucial role in mechanotransduction of fetal rat lung cells (Liu et al., 1994) with specified structural alterations and gating dynamics (Martinac, 2004).
Gene expression is globally mapped under physiologically-mimicking stretch and a number of mammalian and plant genes reacting to mechanical stretch are identified. Different mechanosensitive genes are defined for the metabolism of chondrosarcoma cells exposed to continuous cyclic stretch (Karjalainen et al., 2003) or for the biochemically-induced osteogenesis and bone nodule formation of human ESCs exposed to intermittent cyclic stretch (Li et al., 2013a). In addition to obtaining a candidate list of the differential genes, integrative knowledge of proteins encoded by mechanosensitive genes and of their interactions with putative partners uncovers the programming of genes functionally involved in paracrine signaling of angiogenesis for bladder SMCs subjected to cyclic stretch (Yang et al., 2008). Nowadays, it is also able to profile plant transcriptomes and compare the genes of interest under mechanical stretch. The first expression sequence tags of tension wood are produced from Populus tremula × P. tremuloides tension wood cDNA library (Sterky et al., 2004) and four different cDNA libraries are then constructed from both tensile and opposite wood of bent poplars to identify the highly expressed genes from analyzing the tags in the libraries (Dejardin et al., 2004).
Differential proteins under mechanical stretch are also profiled, especially for those events undetectable in transcriptomic analysis. Cyclic biaxial stretch initiates multiple phosphorylations associated with the newly-identified mechanosensitive proteins in chondrosarcoma cells (Piltti et al., 2008) and promotes 194 and 177 differential proteins related to ECM production, intracellular signaling, cytoskeletal remodeling, and inflammatory response from tenocytes cultured on polyglycolic acid long fibers, respectively (Jiang et al., 2011). Cyclic equiaxial stretch uncovers a few consistent major proteins (TGF-β1, TNF, CASP3, and p53) to hub at the center of the interacting network with the newly-profiled proteins of interest (BAG5, NO66, and eIF-5A) in activated lamina cribrosa cells (Rogers et al., 2012), recapitulating the importance of MAPK and TGF-β signaling pathways in mechanotransduction. Biomechanical and biochemical regulations are also coupled together in stretch-induced proteins, as seen that the TGF-β1-activated up-regulation of α3β1 integrin and uniaxial stretch-induced increase of calponin 3 protein are different from the synergistic up-regulation of calponin 1 gene for human MSCs (Kurpinski et al., 2009). In plant, 5 and 12 proteins are specified in the differentiating tissue of tension-induced wood in Eucalyptus camaldulensisL. (Baba et al., 2000) and in the tensile wood of Eucalyptus gunnii associated with growth strain (Plomion et al., 2003), respectively. Although the available data for plants is much less than those for mammals, such the mechanically-induced proteomic analysis broadens the mechanotransductive extents in plant sciences.
Mechanical compression
Cells are able to sense and respond to internal or external compression. Articular cartilage is a hydrated soft tissue for bearing diarthrodial joints and therefore serves as the main target in compression-induced mechanotransduction. Continuous hydrostatic pressure induces stress-associated transcription factors in primary and immortalized chondrocytes, presumably resulting from the stabilization but not the synthesis of HSP70 mRNA (Kaarniranta et al., 2003). Both dynamic compression and insulin-like growth factor I (IGF-I) regulate, via distinct activating pathways, the metabolic activity of articular chondrocytes in a way that dynamic compression accelerates the biosynthetic response to IGF-I and increases transport of IGF-I into the articular cartilage matrix (Bonassar et al., 2001). Tumor is another typical target mainly due to its uncontrolled growth in a confined space. Tumor cells often experience mechanical compressive stress that stimulates adhesion and migration by a subset of “leader cells” (Tse et al., 2012) whereas tibial compression inhibits the growth and osteolysis of secondary breast tumors (Lynch et al., 2013). These studies provide cues for the potential signaling pathways in response to mechanical compression by these mechanosensitive effectors.
Differential gene expression under mechanical compression is one of key issues in chondrocyte mechanotransduction. Chondrosarcoma cells are more likely sensitive to continuous pressure with several induced genes than cyclic and static pressure with few gene changes under different compression regimens and parameters (Sironen et al., 2000). Not only the hydrostatic pressure regulates the gene expression but it also manipulates the mRNA stability of chondrosarcoma cells, since such immediate-early genes as c-jun, jun-B, and c-myc become up-regulated but destabilized under pressure treatment (Sironen et al., 2002).
Proteomic studies of mechanical compression are applied to understand how different proteins are orchestrated to respond. Proteomic analysis between normal and fatigue axial compressive loads for ulna yields 42 differential proteins encoded by 21 genes that produce an interaction sub-network for differentially expressed proteins (mainly for Raf1 and PDCD8) (Li et al., 2011). Co-culture of osteoblasts-osteocytes exposed to cyclic compression induces the protein release of MMP-3 and -13, inhibits the mRNA expression of Col-II and aggrecan, and promotes 14-3-3ε as a new soluble mediator between subchondral bone and cartilage in osteoarthritis, implying the interactive communications among different types of bone cells (Priam et al., 2013). Moreover, cyclic compression also induces differential productions of typical proteins (aggrecan, Col-I/II, and proteoglycan) for bone marrow-derived mesenchymal progenitor cells (Angele et al., 2004).
Other mechanical stimuli
There are numerous other mechanical stimuli existing in physiology. Combination of both transcriptomic/proteomic analyses and mechanical loading is also important in those mechanical stimuli. Here are two examples.
Gravitational alterations in space are crucial for astronauts’ bone loss and immune suppression under microgravity, which highly depends on the direct and indirect effects of gravitational changes on the relevant cells observed from space missions or ground-based studies (Zayzafoon et al., 2005; Sun et al., 2008; Li et al., 2010; Feuerecker et al., 2013). While gravity-sensitive genes and proteins are occasionally found to be either up-regulated or down-regulated, there is still lack of systematic studies on gene expression and protein production for various types of cells exposed to different microgravity levels (Nichols et al., 2006). The pioneering works of whole-genome analysis on gravitropic stimulation are performed to delineate the transcriptional response mechanisms of plant growth, where Arabidopsis seedlings and root apices in response to clinostat (90° reorientation) and transient gravitropic reorientation (135° reorientation) at different time points (2–60 min) reveal the induction of complex gene expression patterns as a consequence of the fast and transient transcriptional networks and the gravity-induced genes (Moseyko et al., 2002; Kimbrough et al., 2004). Later, the expression profile of microRNAs and genes of human lymphoblastoid cells on simulation of microgravity effect is conducted after being exposed to a rotating wall bioreactor, in which miR-150, miR-34a, miR-423-5p, miR-22, miR-141, miR-618, and miR-222 are found to change significantly (Mangala et al., 2011). Diamagnetic levitation simulation verifies the delay in the development of Drosophila melanogaster from embryo to adult and the significant changes in the transcriptional profile of immune-, stress-, and temperature-related genes (i.e., HSPs) (Herranz et al., 2012), implying the gravity-dependent organism development.
There is an increasing evidence that cells respond to the topographical substrate with physiologically-mimicking stiffness in morphology, proliferation, and differentiation (Li et al., 2013c), suggesting that mechanical features of substrate also play a role in mechanobiology and alter cell transcriptomics/proteomics. Genetic profile of hMSCs presents the promotion of the topography-induced bone formation on nanoscale pitted surface and raised islands (Dalby et al., 2008) and yields a similar expression of p38 MAPK molecule on two width-varied microgrooves but a differential regulation of increased PDGF and integrin expressions and of enhanced VEGF signaling on either microgroove (Biggs et al., 2008). Quantitative proteomics on different topographies uncovers 21 differential proteins related to cell cytoskeleton, metabolism, signaling, and growth identified for osteoblasts placed on planar and carbon nanotube reinforced hydroxyapatite surface (Xu et al., 2008). Further studies integrate the transcriptomics and proteomics analyses to provide the consistent data for transcripts and proteins of fibroblasts on grooved substrate in regulating chromatin remodeling (e.g., HMGA1) and DNA synthesis (e.g., PCNA) (McNamara et al., 2012). Additionally, ECM/cell stiffness is an intrinsic mechanical stimulus that can also regulate cell phenotype. As compared to those dispersed cells, periosteal cells residing in stiffness-varied regions yield the differential genes and the related proteins in regulating ECMs, suggesting a negative correlation between stiffness and differentiation (Horimizu et al., 2013). Mechanosensor-based targeting of membrane stiffness provokes 13 proteins involved in the differentiation of embryonic muscle cells, including galectin-1, annexin III, RhoGD I, and FAK phosphorylation (Grossi et al., 2011). Evidently, ECM/cell topography and stiffness are associated with transcriptomic/proteomic changes in signaling pathways and cellular functions, as well as in metastatic potential of tumor cells (Pozo et al., 2007; Swaminathan et al., 2011; Huang and Ingber, 2005).
Combined mechanical stimuli
Physiologically, the different types of mechanical stimuli or the variety of mechanical patterns or parameters on the same mechanical stimulus are usually coupled together to modulate the cellular and molecular events. Multiple genes and protein effectors are mediated to respond to the acting stimuli or parameters.
Differential regulation of patterns or parameters on single type of stimuli
Increasing evidences indicate that cells respond to mechanical stimuli in a pattern-dependent manner. For example, laminar shear induces EC alignment along the flow direction without initiating cell proliferation whereas turbulent shear stimulates endothelial DNA synthesis in the absence of cell alignment (Davies et al., 1986). Exposure of fibroblasts to oscillatory flow does not promote the development of F-actin stress fibers while the actin polymerization and actin stress fiber formation are fostered under steady flow (Malone et al., 2007). Meanwhile, equi-biaxial stretch inhibits lamellipodia formation via deactivation of Rac signaling whereas uniaxial stretch suppresses lamellipodia along lengthened sides but increases at adjacent ends (Katsumi et al., 2002). Cyclic pressure enhances the aggrecan mRNA expression while static pressure reduces the aggrecan level in primary chondrocytes (Lammi et al., 1994). Moreover, cells respond to mechanical stimuli in a parameter-dependent manner at a given pattern of specific type. Under oscillatory flow, COX-2, RANKL, and OPG mRNA expressions in osteocytes are sensitive to the combination of peak shear stress (0.5, 1, 2, and 5 Pa), oscillatory frequency (0.5, 1, and 2 Hz), and action duration (1, 2, and 4 h) (Li et al., 2012). Continuous tension at 3%–9% strain for 10 days inhibits MSC differentiation while intermittent tension in minutes or hours per day promotes MSC differentiation via Runx2 expression and MAPK signaling (Ward et al., 2007; Shi et al., 2011). These data illustrates that cellular responses are highly sensitive to the patterns and/or parameters of mechanical stimuli that are imposed.
Combinatory impacts of different types of stimuli
Cells usually undergo in vivo distinct types of mechanical loads that are inextricably coupled and are put into effect synergistically. For example, biomechanical tests of torsion-tension of cadaveric femurs reveals the simultaneous, coupled impacts of mechanical torsion and tension (Zdero et al., 2011). Tensile strain amplification from tissue to cellular level in bone is induced by fluid drag forces on bone cells (You et al., 2001), implying the cross-talk between tensile stretch and shear flow. When an equi-biaxial tension and a steady flow are applied separately to fibroblasts in the presence or absence of RhoA inhibitor, the mechanical stimuli regulate fibronectin reorganization and recruitment in different ways (Steward et al., 2011). Obviously, one type of mechanical load alone is unlikely sufficient to generate the required mechanotransductive signaling events for a specific function phenotype. An example to echo this point is the combinatory regulations of surface shear (±25° oscillation at 1 Hz) and cyclic axial compression (0.4 mm amplitude or 10%–20% strain at 1 Hz) for chondrogenic differentiation of hBMSCs. Here the combination enables to mediate chondrogenic gene expression and sulphated glycosaminoglycan and Col-II deposit but either alone does not work (Schatti et al., 2011). Moreover, chondrosarcoma cells yield a less profound effect on the gene expression profile under hydrostatic pressure (continuous and 0.5 Hz cyclic pressure at 5 MPa) than that under cyclic stretch (8% strain at 0.5 Hz) (Karjalainen et al., 2003).
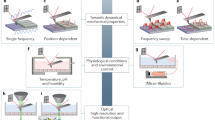
To date, cell mechanotransduction has attracted much attentions to understand, from both theoretical and experimental aspects, the mechanisms how a cell adapts the mechanical stimuli and how the mechanical microenvironments are remodeled. The progress in combining mechanobiology with omics approaches also demonstrates that omics analysis is efficient in screening mechanosensitive genes or proteins in various mechanical stimuli (Fig. 2).
Conceptual demonstration of mechanome to illustrate the combination of mechanobiology/mechanochemistry and genome/transcriptome/proteome. Different mechanical stimuli mediate distinct responsive functions at molecule, cell, or tissue level, while the omics techniques map the entire sets of molecular events of an organism. The combined field helps to uncover globally the mysteries of mechanobiology and mechanochemistry from the viewpoint of omics analyses at different levels
Progresses and Perspective of Mechanomics
Omics approaches, such as transcriptomics and proteomics, are powerful system-level tools that can be applied not only in biology and medicine but also in mechanobiology. However, no system-level omics definition is proposed to summarize the roles of mechanical forces in biological processes, i.e., mechanome. Mapping the mechanome of cells furthers the understandings of mechanosensation and mechanotransduction. Brief discussion is conducted here since only few papers are currently published on this special topic.
Concept of mechanomics
The definition of mechanome is originated for globally elucidating the functions of mechanical forces existing in vivo at cellular and molecular scales (Lang, 2007; Song et al., 2012, 2013). It is then extended to describe the events in tissue, organ, or even whole organism, aiming to understand ultimately the roles of mechanical stress from nano- and micro- to macro-scopic viewpoints (Song et al., 2012). It is necessary to collect the information of mechanical force distribution at different levels, and map the cross-talks between mechanical forces and biological functions. This turns out to a new field, mechanomics. The term mechanomics was first proposed for combining the nuclear magnetic resonance technique with bioinformatics strategies to characterize the protein-ligand interactions across large families of proteins (Sem et al., 2001). Evidently, such definition is distinct with what we discuss here. Regardless of this, the term can be borrowed for describing the mechanically-induced events at multiple levels. Fig. 2 demonstrates the conceptual flowchart of this emerging field. It seeks to understand the fundamental mechanical or physical processes that are common to biological function and to study how forces are transmitted and transduced to regulate biological responses. A similar term of physicomics was also proposed to deal with physical parameters such as pressure, temperature, electromagnetic fields etc. that are involved in cell, tissue, or physiology (van Loon, 2009). To date, quite a few works from the viewpoint of mechanomics are found on transcriptomic and proteomic analyses for the combined mechanical stimuli. Mapping the global responses is critical since multiple mechanical stimuli are associated with the multiple cellular and molecular events (Fig. 2).
Limited progresses in the fields related to mechanomics
Since the term mechanomics is proposed from a distinct viewpoint of drug design (Sem et al., 2001), it has been submerged away from the community of mechanobiology for a while. To date, only a few works are reported along the line described here. One example is that understanding the roles of mechanical forces and machinery (e.g., biological motors and polymerization of filament) opens the window to new strategies for molecular medicine (Wang et al., 2008; Huang et al., 2013). Two more examples are to map the mechanome of live stem cells using both fluorescent microbeads and computational fluid dynamics (CFD) simulation and to measure local strain fields in situ at the fluid-cell interface, which provides mechanistic insights into the roles of mechanical forces in lineage commitment as it unfolds (Wang et al., 2008; Song et al., 2012, 2013). In fact, a body of works, such as those aforementioned, have already adopted the idea or concept of mechanomics to elucidate the cellular and molecular events on different mechanical stimuli even though the term mechanomics is not used explicitly.
Biomechanical and biological approaches appropriate for mechanomics
Biomechanical assays and techniques developed previously are able to be applied in mechanomics. Shear flow, tensile stretch, and mechanical compression are most frequently tested using in-house developed or commercial instruments such as parallel flow chamber, membrane-stretch apparatus, or osmotic/hydrostatic compression device. As indicated in the literatures (van Loon, 2009; Long et al., 2011), parallel or disk flow chamber is used to mimic physiological blood or interstitial flow, uni-, bi-, or equi-axial stretch apparatus is applied to initiate the substrate or ECM tension, and tubular or spheroidal compression device is employed to represent physiological compression. In addition to these approaches, micropipette suction assay applies forces on the cell or vacuole membrane by deforming them (Huang et al., 2004; Fu et al., 2011b). Optical tweezers utilize an optical gradient to trap and exert forces on refractive microbeads (Zhang et al., 2008; Sun et al., 2009; Li et al., 2013b) and magnetic tweezers apply twisting forces via a magnetic field to firmly-attached magnetic microbeads on cell surface (Grossi et al., 2011). Atomic force microscopy enables to quantify rupture force/bond lifetime of protein-protein interactions at single molecule level (Lü et al., 2006). Besides, microgravity simulators such as clinostat, rotating-wall bioreactor, random positioning machine, or magnetic levitation are considered to stimulate the space microgravity effects for ground-based species while hypergravity centrifuges are useful tools for mimicking the hypergravity impacts in space life sciences. Micro-patterned or micro-fabricated substrates are assumed to mimic the physiological topography of microenvironment or niche and then applied in mechanobiology studies of cell proliferation and differentiation (Li et al., 2013c). Meanwhile, coupling the typical fluorescent assays such as fluorescence resonance energy transfer with those mechanical assays also enables to visualize the intracellular in situ events under given mechanical stimuli.
Transcriptomic/proteomic approaches are also critical for the study of mechanomics. Much progress has been made from routine chip-based analysis to the advanced RNA-seq techniques (e.g., SAGE) or from conventional 2-D gel assay to the hi-tech proteomic techniques (e.g., iTRAQ, and LC-MS/MS).
Future perspectives of mechanomics
Mechanomics is a young but fast developing field. Promising clues are emerging in its related aspects in recent years. The framework is exemplified in Fig. 2 to clarify how all these related fields can be linked together. While there should be lots of works to pursue, several potential issues are proposed from the above discussions.
Biological or physiological significance yield the top priority in mechanomic studies. It is little known about how mechanical forces appearing in cell or tissue contribute to development, physiology, and diseases. Molecular mechanisms (e.g., mechanosensors) by which cells sense and respond to mechanical signals is the major challenge we are facing, since we are still far away to establish the responsive network under global mechanical stimuli. More biological models should be considered, as the biologists usually do, to broaden the vision of study for biological diversity. Expression profiling under pathological mechanical conditions is another hotspot in the mechanobiology of tumor growth (Carey et al., 2012).
Replication of the in vivo mechanical patterns or parameters is also a prior challenge to confine the understanding how mechanomics works for cells. In a living organism, the in vivo mechanical environment is quite complicated and usually coupled with other physical and chemical factors. Thus, it is difficult to replicate the local biomechanical environment that surrounds cells or comprises tissues and is hard to determine the physiological mechanical patterns or parameters. Meanwhile, existing in vitro data for cells exposed to mechanical stimuli are usually compared with those under static culture conditions, which require the careful design of control cases that are expected to subtract reasonably background noises from measured signals. More attentions should also be paid for those stimuli not indicated here (e.g., vibration, sound, touch).
Developing new in vitro assays specific for mechanomics is also an important issue. New hi-tech techniques in related fields provide the opportunities for promoting the mechanomics. On-chip strain sensors developed by micro-fabrication combined with mechanical tension promise to test small samples and mechanical loading in parallel, to reduce the labor-consumption, and to enhance testing efficiency (MacQueen et al., 2012). Micro-fabricated composite material screening array is also able to determine the combined effects of substrate stretch, soluble cues, and matrix proteins on small populations of primary cells, which eventually enable us to profile gene expression from a single cell (Moraes et al., 2013). Meanwhile, the state-of-art techniques in biology direct the analyses of cells in situ or in vivo, or carry out immediately the post mortem to retain the features of the native environment. It is then possible to design appropriate in vitro experiments that critically test the most dominant mechanical characteristics existing in vivo. New hi-tech techniques of 2nd-generation sequencing or single-cell sequencing could be efficient candidates for mapping the mechanomics of various species (Lawrie et al., 2008).
Large-scale data mining and bioinformatics analysis are indispensable in this field. To facilitate the integration of database, an open-access and downloadable mechanomics database should be created worldwide, serving as a web-based resource like NCBI that collates quantitative analyses of mechanomics. One key feature of the database is able to map globally the mechanical environment of cells and provide the quantitative information for transcripts and proteins from various sources. Another is the capability of large-scale, elaborated analyses with interactive interface for the users who attempt to mine the existing data or submit newly measured data using various search engines or experimental assays.
Conclusions
Combination of mechanobiology and transcriptomics/proteomics is an emerging field for globally understanding the gene expression and protein production in response to multiple types of mechanical stimuli. Evidently, mechanomics is still in its embryonic stage. With the high-throughput analyses in transcriptomics and proteomics and the state-of-art techniques in biomechanics, one expects to define the required mechanical variables and to provide an integrated profile of signaling events from the viewpoint of mechanome at molecular and cellular levels.
Abbreviations
- 2-D:
-
two dimensional
- ACL:
-
anterior cruciate ligament
- EC:
-
endothelial cell
- ECM:
-
extracellular matrix
- MPC:
-
mesenchymal progenitor cell
- HSP:
-
heat shock protein
- IGF-I:
-
insulin-like growth factor I
- iTRAQ:
-
isobaric tags for relative and absolute quantification
- LC:
-
liquid chromatograph
- MS:
-
mass spectrometry
- MSC:
-
mesenchymal stem cell
- SAGE:
-
serial analysis of gene expression
- SMC:
-
smooth muscle cell
References
Angele P, Schumann D, Angele M, Kinner B, Englert C, Hente R, Fuchtmeier B, Nerlich M, Neumann C, Kujat R (2004) Cyclic, mechanical compression enhances chondrogenesis of mesenchymal progenitor cells in tissue engineering scaffolds. Biorheology 41:335–346
Arnsdorf EJ, Tummala P, Kwon RY, Jacobs CR (2009) Mechanically induced osteogenic differentiation—the role of RhoA, ROCKII and cytoskeletal dynamics. J Cell Sci 122:546–553
Baba K, Asada T, Hayashi T (2000) Relation between developmental changes on anatomical structure and on protein pattern in differentiating xylem of tension wood. J Wood Sci 46:1–7
Barakat AI, Lieu DK (2003) Differential responsiveness of vascular endothelial cells to different types of fluid mechanical shear stress. Cell Biochem Biophys 38:323–343
Biggs MJP, Richards RG, McFarlane S, Wilkinson CDW, Oreffo ROC, Dalby MJ (2008) Adhesion formation of primary human osteoblasts and the functional response of mesenchymal stem cells to 330 nm deep microgrooves. J R Soc Interface 5:1231–1242
Bonassar LJ, Grodzinsky AJ, Frank EH, Davila SG, Bhaktav NR, Trippel SB (2001) The effect of dynamic compression on the response of articular cartilage to insulin-like growth factor-I. J Orthop Res 19:11–17
Brooks AR, Lelkes PI, Rubanyi GM (2002) Gene expression profiling of human aortic endothelial cells exposed to disturbed flow and steady laminar flow. Physiol Genomics 9:27–41
Carey SP, D’Alfonso TM, Shin SJ, Reinhart-King CA (2012) Mechanobiology of tumor invasion: engineering meets oncology. Crit Rev Oncol Hematol 83:170–183
Carter DR, Beaupre GS, Giori NJ, Helms JA (1998) Mechanobiology of skeletal regeneration. Clin Orthop 355:S41–S55
Chaplain MAJ, Graziano L, Preziosi L (2006) Mathematical modelling of the loss of tissue compression responsiveness and its role in solid tumour development. Math Med Biol 23:197–229
Chen BPC, Li YS, Zhao YH, Chen KD, Li S, Lao JM, Yuan SL, Shyy JYJ, Chien S (2001) DNA microarray analysis of gene expression in endothelial cells in response to 24-h shear stress. Physiol Genomics 7:55–63
Cohen DM, Chen CS (2008) Mechanical control of stem cell differentiation. In: Bhatia S, Polak J (eds) StemBook. Harvard Stem Cell Institute, Cambridge, pp 1–16
Cui YH, Huo B, Sun SJ, Yang F, Gao YX, Pan J, Long M (2011) Fluid dynamics analysis of a novel micropatterned cell bioreactor. Ann Biomed Eng 39:1592–1605
Dado D, Sagi M, Levenberg S, Zemel A (2012) Mechanical control of stem cell differentiation. Regen Med 7:101–116
Dalby MJ, Andar A, Nag A, Affrossman S, Tare R, McFarlane S, Oreffo ROC (2008) Genomic expression of mesenchymal stem cells to altered nanoscale topographies. J R Soc Interface 5:1055–1065
Davies PF, Remuzzi A, Gordon EJ, Dewey CF, Gimbrone MA (1986) Turbulent fluid shear-stress induces vascular endothelial-cell turnover in vitro. Proc Natl Acad Sci USA 83:2114–2117
Davies PF, Spaan JA, Krams R (2005) Shear stress biology of the endothelium. Ann Biomed Eng 33:1714–1718
Dejardin A, Leple JC, Lesage-Descauses MC, Costa G, Pilate G (2004) Expressed sequence tags from poplar wood tissues—a comparative analysis from multiple libraries. Plant Biol 6:55–64
Evans E, Yeung A (1989) Apparent viscosity and cortical tension of blood granulocytes determined by micropipet aspiration. Biophys J 56:151–160
Feuerecker M, Feuerecker B, Matzel S, Long M, Strewe C, Kaufmann I, Hoerl M, Schelling G, Rehm M, Chouker A (2013) Five days of head-down-tilt bed rest induces noninflammatory shedding of L-selectin. J Appl Physiol 115:235–242
Fu CL, Tong CF, Dong C, Long M (2011a) Modeling of cell aggregation dynamics governed by receptor–ligand binding under shear flow. Cell Mol Bioeng 4:427–441
Fu CL, Tong CF, Wang ML, Gao YX, Zhang Y, Lu SQ, Liang SL, Dong C, Long M (2011b) Determining beta(2)-integrin and intercellular adhesion molecule 1 binding kinetics in tumor cell adhesion to leukocytes and endothelial cells by a gas-driven micropipette assay. J Biol Chem 286:34777–34787
Geiger B, Spatz JP, Bershadsky AD (2009) Environmental sensing through focal adhesions. Nat Rev Mol Cell Biol 10:21–33
Grossi A, Lametsch R, Karlsson AH, Lawson MA (2011) Mechanical stimuli on C2C12 myoblasts affect myoblast differentiation, focal adhesion kinase phosphorylation and galectin-1 expression: a proteomic approach. Cell Biol Int 35:579–586
Herranz R, Larkin OJ, Dijkstra CE, Hill RJ, Anthony P, Davey MR, Eaves L, van Loon JJWA, Medina FJ, Marco R (2012) Microgravity simulation by diamagnetic levitation: effects of a strong gradient magnetic field on the transcriptional profile of Drosophila melanogaster. BMC Genomics 13(1):52
Heydarkhan-Hagvall S, Chien S, Nelander S, Li YC, Yuan SL, Lao JM, Haga JH, Lian I, Nguyen P, Risberg B et al (2006) DNA microarray study on gene expression profiles in co-cultured endothelial and smooth muscle cells in response to 4-and 24-h shear stress. Mol Cell Biochem 281:1–15
Horimizu M, Kawase T, Tanaka T, Okuda K, Nagata M, Burns DM, Yoshie H (2013) Biomechanical evaluation by AFM of cultured human cell-multilayered periosteal sheets. Micron 48:1–10
Huang S, Ingber DE (2005) Cell tension, matrix mechanics, and cancer development. Cancer Cell 8:175–176
Huang J, Chen J, Chesla SE, Yago T, Mehta P, McEver RP, Zhu C, Long M (2004) Quantifying the effects of molecular orientation and length on two-dimensional receptor–ligand binding kinetics. J Biol Chem 279:44915–44923
Huang C, Holfeld J, Schaden W, Orgill D, Ogawa R (2013) Mechanotherapy: revisiting physical therapy and recruiting mechanobiology for a new era in medicine. Trends Mol Med 19:555–564
Ingber DE (1997) Tensegrity: the architectural basis of cellular mechanotransduction. Annu Rev Physiol 59:575–599
Janmey PA, McCulloch CA (2007) Cell mechanics: integrating cell responses to mechanical stimuli. Annu Rev Biomed Eng 9:1–34
Jiang YK, Liu HW, Li H, Wang FJ, Cheng K, Zhou GD, Zhang WJ, Ye ML, Cao YL, Liu W et al (2011) A proteomic analysis of engineered tendon formation under dynamic mechanical loading in vitro. Biomaterials 32:4085–4095
Kaarniranta K, Elo MA, Sironen RK, Karjalainen HM, Helminen HJ, Lammi MJ (2003) Stress responses of mammalian cells to high hydrostatic pressure. Biorheology 40:87–92
Kang YY, Lu SQ, Ren P, Huo B, Long M (2012) Molecular dynamics simulation of shear- and stretch-induced dissociation of P-selectin/PSGL-1 complex. Biophys J 102:112–120
Karjalainen HM, Sironen RK, Elo MA, Kaarniranta K, Takigawa M, Helminen HJ, Lammi MJ (2003) Gene expression profiles in chondrosarcoma cells subjected to cyclic stretching and hydrostatic pressure. A cDNA array study. Biorheology 40:93–100
Katsumi A, Milanini J, Kiosses WB, del Pozo MA, Kaunas R, Chien S, Hahn KM, Schwartz MA (2002) Effects of cell tension on the small GTPase Rac. J Cell Biol 158:153–164
Katsumi A, Orr AW, Tzima E, Schwartz MA (2004) Integrins in mechanotransduction. J Biol Chem 279:12001–12004
Kimbrough JM, Salinas-Mondragon R, Boss WF, Brown CS, Sederoff HW (2004) The fast and transient transcriptional network of gravity and mechanical stimulation in the Arabidopsis root apex. Plant Physiol 136:2790–2805
Kurpinski K, Chu J, Wang DJ, Li S (2009) Proteomic profiling of mesenchymal stem cell responses to mechanical strain and TGF-beta 1. Cell Mol Bioeng 2:606–614
Lammi MJ, Inkinen R, Parkkinen JJ, Hakkinen T, Jortikka M, Nelimarkka LO, Jarvelainen HT, Tammi MI (1994) Expression of reduced amounts of structurally altered aggrecan in articular-cartilage chondrocytes exposed to high hydrostatic-pressure. Biochem J 304:723–730
Lang M (2007) Lighting up the mechanome. Bridge 37:11–16
Lawrie CH, Gal S, Dunlop HM, Pushkaran B, Liggins AP, Pulford K, Banham AH, Pezzella F, Boultwood J, Wainscoat JS et al (2008) Detection of elevated levels of tumour-associated microRNAs in serum of patients with diffuse large B-cell lymphoma. Br J Haematol 141:672–675
Li T (2008) A mechanics model of microtubule buckling in living cells. J Biomech 41:1722–1729
Li YSJ, Haga JH, Chien S (2005) Molecular basis of the effects of shear stress on vascular endothelial cells. J Biomech 38:1949–1971
Li H, Chen JA, Zhang Y, Sun SJ, Tao ZL, Long M (2010) Effects of oriented substrates on cell morphology, the cell cycle, and the cytoskeleton in Ros 17/2.8 cells. Sci Chin Life Sci 53:1085–1091
Li JL, Zhang F, Chen JY (2011) An integrated proteomics analysis of bone tissues in response to mechanical stimulation. BMC Syst Biol 5:S7
Li J, Rose E, Frances D, Sun Y, You LD (2012) Effect of oscillating fluid flow stimulation on osteocyte mRNA expression. J Biomech 45:247–251
Li M, Li X, Meikle MC, Islam I, Cao T (2013a) Short periods of cyclic mechanical strain enhance triple-supplement directed osteogenesis and bone nodule formation by human embryonic stem cells in vitro. Tissue Eng Part A 19:2130–2137
Li N, Mao D, Lu S, Tong C, Zhang Y, Long M (2013b) Distinct binding affinities of Mac-1 and LFA-1 in neutrophil activation. J Immunol 190:4371–4381
Li Z, Gong YW, Sun SJ, Du Y, Lu DY, Liu XF, Long M (2013c) Differential regulation of stiffness, topography, and dimension of substrates in rat mesenchymal stem cells. Biomaterials 34:7616–7625
Liang S, Fu C, Wagner D, Guo H, Zhan D, Dong C, Long M (2008) Two-dimensional kinetics of beta(2)-integrin and ICAM-1 bindings between neutrophils and melanoma cells in a shear flow. Am J Physiol 294:C1604–C1605
Liu MY, Xu J, Tanswell AK, Post M (1994) Inhibition of mechanical strain-induced fetal-rat lung-cell proliferation by gadolinium, a stretch-activated channel blocker. J Cell Physiol 161:501–507
Long M, Sato M, Lim CT, Wu JH, Adachi T, Inoue Y (2011) Advances in experiments and modeling in micro- and nano-biomechanics: a mini review. Cell Mol Bioeng 4:327–339
Lü SQ, Ye ZY, Zhu C, Long M (2006) Quantifying the effects of contact duration, loading rate, and approach velocity on P-selectin–PSGL-1 interactions using AFM. Polymer 47:2539–2547
Lü DY, Liu XF, Gao YX, Huo B, Kang YY, Chen J, Sun SJ, Chen L, Luo XD, Long M (2013) Asymmetric migration of human keratinocytes under mechanical stretch and cocultured fibroblasts in a wound repair model. PLoS ONE 8:e74563
Lynch ME, Brooks D, Mohanan S, Lee MJ, Polamraju P, Dent K, Bonassar LJ, van der Meulen MCH, Fischbach C (2013) In vivo tibial compression decreases osteolysis and tumor formation in a human metastatic breast cancer model. J Bone Miner Res 28:2357–2367
MacQueen L, Chebotarev O, Simmons CA, Sun Y (2012) Miniaturized platform with on-chip strain sensors for compression testing of arrayed materials. Lab Chip 12:4178–4184
Malone AMD, Batra NN, Shivaram G, Kwon RY, You LD, Kim CH, Rodriguez J, Jair K, Jacobs CR (2007) The role of actin cytoskeleton in oscillatory fluid flow-induced signaling in MC3T3-E1 osteoblasts. Am J Physiol 292:C1830–C1836
Mangala LS, Zhang Y, He Z, Emami K, Ramesh GT, Story M, Rohde LH, Wu H (2011) Effects of simulated microgravity on expression profile of microRNA in human lymphoblastoid cells. J Biol Chem 286:32483–32490
Martinac B (2004) Mechanosensitive ion channels: molecules of mechanotransduction. J Cell Sci 117:2449–2460
McNamara LE, Burchmore R, Riehle MO, Herzyk P, Biggs MJ, Wilkinson CD, Curtis AS, Dalby MJ (2012) The role of microtopography in cellular mechanotransduction. Biomaterials 33:2835–2847
Moraes C, Likhitpanichkul M, Lam CJ, Beca BM, Sun Y, Simmons CA (2013) Microdevice array-based identification of distinct mechanobiological response profiles in layer-specific valve interstitial cells. Integr Biol 5:673–680
Moseyko N, Zhu T, Chang HS, Wang X, Feldman LJ (2002) Transcription profiling of the early gravitropic response in Arabidopsis using high-density oligonucleotide probe microarrays. Plant Physiol 130:720–728
Murrell M, Pontani LL, Guevorkian K, Cuvelier D, Nassoy P, Sykes C (2011) Spreading dynamics of biomimetic actin cortices. Biophys J 100:1400–1409
Nichols HL, Zhang N, Wen XJ (2006) Proteomics and genomics of microgravity. Physiol Genomics 26:163–171
Piltti J, Hayrinen J, Karjalainen HM, Lammi MJ (2008) Proteomics of chondrocytes with special reference to phosphorylation changes of proteins in stretched human chondrosarcoma cells. Biorheology 45:323–335
Plomion C, Pionneau C, Bailleres H (2003) Analysis of protein expression along the normal to tension wood gradient in Eucalyptus gunnii. Holzforschung 57:353–358
Porat Z, Yaron I, Katz BZ, Kam Z, Geiger B (2011) Shear flow-induced formation of tubular cell protrusions in multiple myeloma cells. J Cell Physiol 226:3197–3207
Pozo L, Sanchez-Carrillo JJ, Martinez A, Blanes A, Diaz-Cano SJ (2007) Differential kinetic features by tumour topography in cutaneous small-cell neuroendocrine (Merkel cell) carcinomas. J Eur Acad Dermatol 21:1220–1228
Priam S, Bougault C, Houard X, Gosset M, Salvat C, Berenbaum F, Jacques C (2013) Identification of soluble 14-3-3epsilon as a new subchondral bone mediator involved in cartilage degradation in osteoarthritis. Arthritis Rheumatol 65:1831–1842
Qi YX, Jiang J, Jiang XH, Wang XD, Ji SY, Han Y, Long DK, Shen BR, Yan ZQ, Chien S et al (2011) PDGF-BB and TGF-beta 1 on cross-talk between endothelial and smooth muscle cells in vascular remodeling induced by low shear stress. Proc Natl Acad Sci USA 108:1908–1913
Richard MN, Deniset JF, Kneesh AL, Blackwood D, Pierce GN (2007) Mechanical stretching stimulates smooth muscle cell growth, nuclear protein import, and nuclear pore expression through mitogen-activated protein kinase activation. J Biol Chem 282:23081–23088
Rogers RS, Dharsee M, Ackloo S, Sivak JM, Flanagan JG (2012) Proteomics analyses of human optic nerve head astrocytes following biomechanical strain. Mol Cell Proteomics 11(M111):012302
Schatti O, Grad S, Goldhahn J, Salzmann G, Li Z, Alini M, Stoddart MJ (2011) A combination of shear and dynamic compression leads to mechanically induced chondrogenesis of human mesenchymal stem cells. Eur Cells Mater 22:214–225
Sem DS, Yu L, Coutts SM, Jack R (2001) Object-oriented approach to drug design enabled by NMR SOLVE: first real-time structural tool for characterizing protein-ligand interactions. J Cell Biochem 84:99–105
Shi Y, Li HW, Zhang XL, Fu YJ, Huang Y, Lui PPY, Tang TT, Dai KR (2011) Continuous cyclic mechanical tension inhibited Runx2 expression in mesenchymal stem cells through RhoA-ERK1/2 pathway. J Cell Physiol 226:2159–2169
Shieh AC, Rozansky HA, Hinz B, Swartz MA (2011) Tumor cell invasion is promoted by interstitial flow-induced matrix priming by stromal fibroblasts. Cancer Res 71:790–800
Silkworth JB, Stehbens WE (1975) Shape of endothelial cells in en face preparations of rabbit blood-vessels. Angiology 26:474–487
Sironen R, Elo M, Kaarniranta K, Helminen HJ, Lammi MJ (2000) Transcriptional activation in chondrocytes submitted to hydrostatic pressure. Biorheology 37:85–93
Sironen RK, Karjalainen HM, Torronen K, Elo MA, Kaarniranta K, Takigawa M, Helminen HJ, Lammi MJ (2002) High pressure effects on cellular expression profile and mRNA stability. A cDNA array analysis. Biorheology 39:111–117
Soltz MA, Ateshian GA (2000) Interstitial fluid pressurization during confined compression cyclical loading of articular cartilage. Ann Biomed Eng 28:150–159
Song MJ, Brady-Kalnay SM, McBride SH, Phillips-Mason P, Dean D, Tate MLK (2012) Mapping the mechanome of live stem cells using a novel method to measure local strain fields in situ at the fluid-cell interface. PLoS ONE 7:e43601
Song MJ, Dean D, Knothe Tate ML (2013) Mechanical modulation of nascent stem cell lineage commitment in tissue engineering scaffolds. Biomaterials 34:5766–5775
Stamenovic D, Ingber DE (2002) Models of cytoskeletal mechanics of adherent cells. Biomech Model Mechanobiol 1:95–108
Sterky F, Bhalerao RR, Unneberg P, Segerman B, Nilsson P, Brunner AM, Charbonnel-Campaa L, Lindvall JJ, Tandre K, Strauss SH et al (2004) A populus EST resource for plant functional genomics. Proc Natl Acad Sci USA 101:13951–13956
Steward RL, Cheng CM, Ye JD, Bellin RM, LeDuc PR (2011) Mechanical stretch and shear flow induced reorganization and recruitment of fibronectin in fibroblasts. Sci Rep 1:147
Sun SJ, Gao YX, Shu NJ, Tang ZM, Tao ZL, Long M (2008) A novel counter sheet-flow sandwich cell culture device for mammalian cell growth in space. Microgravity Sci Technol 20:115–120
Sun GY, Zhang Y, Huo B, Long M (2009) Parametric analysis for monitoring 2D kinetics of receptor–ligand binding. Cell Mol Bioeng 2:495–503
Swaminathan V, Mythreye K, O’Brien ET, Berchuck A, Blobe GC, Superfine R (2011) Mechanical stiffness grades metastatic potential in patient tumor cells and in cancer cell lines. Cancer Res 71:5075–5080
Tamura I, Rosenbloom J, Macarak E, Chaqour B (2001) Regulation of Cyr61 gene expression by mechanical stretch through multiple signaling pathways. Am J Physiol 281:C1524–C1532
Tan SD, de Vries TJ, Kuijpers-Jagtman AM, Semeins CM, Elverts V, Klein-Nulend J (2007) Osteocytes subjected to fluid flow inhibit osteoclast formation and bone resorption. Bone 41:745–751
Tse JM, Cheng G, Tyrrell JA, Wilcox-Adelman SA, Boucher Y, Jain RK, Munn LL (2012) Mechanical compression drives cancer cells toward invasive phenotype. Proc Natl Acad Sci USA 109:911–916
van Loon JJWA (2009) Mechanomics and physicomics in gravisensing. Microgravity Sci Technol 21:159–167
Wang JHC (2006) Mechanobiology of tendon. J Biomech 39:1563–1582
Wang YX, Shyy JYJ, Chien S (2008) Fluorescence proteins, live-cell imaging, and mechanobiology: seeing is believing. Annu Rev Biomed Eng 10:1–38
Wang XL, Fu A, Spiro C, Lee HC (2009) Proteomic analysis of vascular endothelial cells-effects of laminar shear stress and high glucose. J Proteomics Bioinform 2:445–454
Ward DF, Salasznyk RM, Klees RF, Backiel J, Agius P, Bennett K, Boskey A, Plopper GE (2007) Mechanical strain enhances extracellular matrix-induced gene focusing and promotes osteogenic differentiation of human mesenchymal stem cells through an extracellular-related kinase-dependent pathway. Stem Cells Dev 16:467–479
Wirtz D, Konstantopoulos K, Searson PC (2011) The physics of cancer: the role of physical interactions and mechanical forces in metastasis. Nat Rev Cancer 11:512–522
Wright MO, Nishida K, Bavington C, Godolphin JL, Dunne E, Walmsley S, Jobanputra P, Nuki G, Salter DM (1997) Hyperpolarisation of cultured human chondrocytes following cyclical pressure-induced strain: evidence of a role for alpha 5 beta 1 integrin as a chondrocyte mechanoreceptor. J Orthop Res 15:742–747
Xu J, Khor KA, Sui J, Zhang J, Tan TL, Chen WN (2008) Comparative proteomics profile of osteoblasts cultured on dissimilar hydroxyapatite biomaterials: an iTRAQ-coupled 2-D LC–MS/MS analysis. Proteomics 8:4249–4258
Yang R, Amir J, Liu HB, Chaqour B (2008) Mechanical strain activates a program of genes functionally involved in paracrine signaling of angiogenesis. Physiol Genomics 36:1–14
Yang F, Gao YX, Zhang Y, Chen J, Long M (2009) Developing a microfluidic-based system to quantify cell capture efficiency. Sci China Ser C 52:173–181
Yi W, Sun Y, Wei XF, Gu CH, Dong XC, Kang XJ, Guo SZ, Dou KF (2010) Proteomic profiling of human bone marrow mesenchymal stem cells under shear stress. Mol Cell Biochem 341:9–16
You LD, Cowin SC, Schaffler MB, Weinbaum S (2001) A model for strain amplification in the actin cytoskeleton of osteocytes due to fluid drag on pericellular matrix. J Biomech 34:1375–1386
Zayzafoon M, Meyers VE, McDonald JM (2005) Microgravity: the immune response and bone. Immunol Rev 208:267–280
Zdero R, McConnell AJ, Peskun C, Syed KA, Schemitsch EH (2011) Biomechanical measurements of torsion–tension coupling in human cadaveric femurs. J Biomech Eng 133:014501-1–014501-6
Zhan DY, Zhang Y, Long M (2012) Spreading of human neutrophils on an ICAM-1-immobilized substrate under shear flow. Chin Sci Bull 57:769–775
Zhang Y, Sun GY, Lu SQ, Li N, Long M (2008) Low spring constant regulates P-selectin–PSGL-1 bond rupture. Biophys J 95:5439–5448
Zhou D, Lee HS, Villarreal F, Teng A, Lu E, Reynolds S, Qin C, Smith J, Sung KLP (2005) Differential MMP-2 activity of ligament cells under mechanical stretch injury: an in vitro study on human ACL and MCL fibroblasts. J Orthop Res 23:949–957
Acknowledgements
This work was supported by National Basic Research Program (973 Program) (No. 2011CB710904), National Natural Science Foundation (No. 31110103918), Strategic Priority Research Program grants XDA01030102 and XDA04020219, and National High Technology Research and Development Program grant 2011AA020109, and Knowledge Innovation Program grant KJCX2-YW-L08.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
About this article
Cite this article
Wang, J., Lü, D., Mao, D. et al. Mechanomics: an emerging field between biology and biomechanics. Protein Cell 5, 518–531 (2014). https://doi.org/10.1007/s13238-014-0057-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13238-014-0057-9