Abstract
Most fungi display a mixed mating system with both asexual and sexual reproduction. The timing of the two modes of reproduction must be carefully coordinated through signal perception and coordination in the cell along with chromatin modification. Here, we investigated coordination of reproductive output by investigating the function of the histone chaperone anti-silencing factor 1 (ASF1) in a fungal species amenable to characterization of both asexual and sexual reproduction. We used knockout approach to show that SeASF1 influenced asexual and sexual reproduction in Stemphylium eturmiunum. SeASF1-deleted strains failed to produce pseudothecia, but produce abnormal conidia and showed an irregular distribution of nuclei in mycelium. Transcriptome sequencing was then used to identify genes with altered expression in the SeASF1-deleted strains. The transcriptional expression of the identified SeDJ-1 was strongly regulated by SeASF1. The interaction of SeDJ-1 and SeASF1 was confirmed using Y2H, Co-IP, and pull-down. Due to some components of phosphatidylinositol 3-kinase/protein kinase B (PI3K/AKT) signaling pathway were known to interact with DJ-1 in mammals, we verified SePI3K, an element of PI3K/AKT signaling pathway in S. eturmiunum, was directly linked to SeDJ-1 and then these two proteins were defined as a coordinator of reproduction. However, knockout of SeDJ-1 or SePI3K altered the asexual and sexual reproduction, but SePI3K recovered the asexual and sexual development of ∆Sedj-1. The SeDJ-1-M6 segment of SeDJ-1 was essential for its interaction with SePI3K and played a critical role in restoring sexual reproduction in the ∆Sepi3k, providing a deep understanding of the regulatory mechanism of SeDJ-1 in S. eturmiunum development. Summarily, SeASF1 is able to trigger SeDJ-1 and SeDJ-1can also activate SePI3K, which is orchestrally involved in asexual and sexual reproduction in S. eturmiunum. All these results reveal that SeASF1 manipulates asexual and sexual reproduction in S. eturmiunum by SeDJ-1 perception of PI3K/AKT signaling pathway. These data highlight the deep similarities in coordinating asexual and sexual processes in both fungi and eukaryotes in general.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Fungi are widely distributed in ecosystems and have a variety of forms, including yeasts, molds, and macro fungi (e.g., mushrooms) (Yuan et al. 2020). In the last 20 years, fungal surveys in Asian countries such as China and Thailand have provided additional evidence of the size of the fungal kingdom (Dai et al. 2015; Hyde et al. 2018). Many fungi species can reproduce both asexually and sexually (holomorph). However, no less than 20% of fungi, especially most of species in ascomycete fungi lack of a known sexual phase (teleomorph) and exclusively reproduce asexually (anamorph) (Dyer and O’Gorman 2011). Studies have shown that amorphic fungi are also likely to reproduce sexually under specific conditions (Han et al. 2003; Kück and Pöggeler 2009). Reproduction is the critical phase distinguishing the winners and losers in the survival of the fittest. Both mitosis and meiosis are deep rooted in the evolution of life (Wilkins and Holliday 2009), and the sexual process shows deep similarity in all eukaryotes, yet details of how gametes are formed and manage to fuse show ample variation (Mori et al. 2015; Nair 2020). Fungi are excellent models to study the evolution and diversification of the sexual process due to the myriad ways in which they have diversified the sexual and asexual process (Fu et al. 2019). It has been suggested that sex is indispensable in fungi (Ni et al. 2011), and although not apparent, sexual reproduction can be detected in the form of recombination. Many species in Ascomycota have a mixed mating system where in sex is reserved for particular occasions and may occur very rarely. The regulatory overlap between asexual and sexual reproduction in fungi is poorly understood but is essential for understanding how environmental signals lead to differential modes of reproduction in fungi or Ascomycota which mostly show a mixed mating system.
The genus Stemphylium is classified into the family Pleosporaceae of the phylum Ascomycota (Câmara et al. 2002; Simmons 1989). Stemphylium is closely related to genera Alternaria and Ulocladium (Simmons 1967). While the sexual state of the asexual Ulocladium has not yet been identified (Wang et al. 2017), the sexual states of some asexual Stemphylium and Alternaria species are revealed as Pleospora and Lewia, respectively (Lucas and Webster 1964; Simmons 1969, 1989; Inderbitzin et al. 2005). Nevertheless, most species within these three genera are mainly classified upon asexual states (Câmara et al. 2002; Woudenberg et al. 2013, 2017). Due to lack of understanding of the sexual mechanism, efforts to link the asexual and sexual states in all these fungi have been challenged. Previous work on S. eturmiunum has shown how it can serve as a model for understanding the interplay between asexual and sexual reproductive modes (Wang et al. 2022).
Sexual reproduction is the predominant reproductive strategy among eukaryotes (Dacks and Roger 1999; Ramesh et al. 2005). In fungi, it depends not only on environmental factors such as habitat, nutrient, light and temperature (Han et al. 2003; Lee et al. 2010; Peberdy 1980; Wang et al. 2016; Wallen and Perlin 2018), but also on their sexual compatibility which is regulated by mating-type genes (Böhm et al. 2013; Coppin et al. 1997). The mating-type genes manipulate selfing and control how cells signal their compatibility through the expression and reception of pheromone components (Bobrowicz et al. 2002; Lin et al. 2011), G proteins (Li et al. 2007; Studt et al. 2013), and velvet proteins (Bayram and Braus 2012). The signaling pathways of mitogen-activated protein kinase (MAPK) (Bayram et al. 2012; Chen et al. 2002; Saito 2010), cell wall integrity (CWI) (Teichert et al. 2014; Zhang et al. 2020), and cyclic adenosine monophosphate/protein kinase A (cAMP-PKA) (Dos Reis et al. 2019) are all highly conserved in eukaryotes and also regulators of sexual mating in fungi. However, many aspects of this regulation remain unclear and sexuality in fungi is a confusing and an interesting topic in biology of many fungi.
There are few genes other than the mating type genes that are known to effect on sexual development. ASF1, anti-silencing function 1, was one such gene which was identified as playing a conserved role in mating in diverse filamentous ascomycetes (Gesing et al. 2012). ASF1, an H3-H4 chaperone are highly conserved from yeast to mammals, was first identified in Saccharomyces cerevisiae (Le et al. 1997). ASF1 plays an important role in nucleosome assembly/disassembly (Avvakumov et al. 2011; Eitoku et al. 2008; Min et al. 2020; Prado et al. 2004; Sanematsu et al. 2006), normal cell cycle progression (Groth et al. 2007; Sutton et al. 2001), genomic instability along with histone modification (Das et al. 2014; Li et al. 2008; Recht et al. 2006), mating type (Le et al. 1997), and sexual reproduction (Gesing et al. 2012; Messiaen et al. 2016).
Because ASF1 plays the multi-faceted roles in the cell and is able to interact with the highly conserved core histone proteins that involved in transforming chromatin between active and inactive states of transcription, we hypothesize that ASF1 is likely to play an important role in regulating the output of asexual and sexual reproduction in the S. eturmiunum, a fungus with a mixed mating system in the Dothideomycetes.
DJ-1 was first identified as a causative gene for autosomal recessive, encoded a causative gene of familial Parkinson’s disease (PARK7) (Bonifati et al. 2003). Recently, evidences show that DJ-1 is an oncogene related to numerous types of cancer (Bai et al. 2012; Chen et al. 2012; Scumaci et al. 2020) and an essential regulator of multiple cellular processes, including anti-oxidative stress, anti-apoptotic effects, and protein degradation (Hijioka et al. 2017; Mukherjee et al. 2015; Taira et al. 2004). As a multifunctional protein, DJ-1 plays a major role in the phosphatidylinositol-3-kinase/protein kinase B (PI3K/AKT) signaling pathway (Yang et al. 2005). The PI3K/AKT signaling pathway manipulates a variety of biological processes, including cell differentiation, proliferation, growth, metabolism, survival, genomic stability, protein synthesis, angiogenesis, cancer development, inhibition of apoptosis, oxidative stress, and regulation of various molecules (Engelman et al. 2006; Gong et al. 2018; Liu et al. 2020; Patra et al. 2019; Srinivasan et al. 2005; Yang et al. 2018; Zhou et al. 2017). This pathway can also modulate sexual reproduction in mammals (Fu et al. 2020; Shao et al. 2019). Manipulation of the expression of this gene may therefore be a means of determining whether the (PI3K/AKT) signaling pathway regulates sexual and asexual reproduction in S. eturmiunum.
ASF1 shows a ubiquitous function for regulating sexual development in animals, plants, and fungi. However, it is not understood how ASF1 may modulate the switch between asexual and sexual development in ascomycete fungi. Here, we show that SeASF1 interacts with SeDJ-1 or SeH4, and that SeDJ-1 as well as SeASF1 promotes the asexual and sexual development in S. eturmiunum. Furthermore, SeDJ-1 also binds to SePI3K and then activates the PI3K/AKT pathway to modulate asexual and sexual features. Thus, ASF1 manipulates asexual and sexual reproduction in S. eturmiunum by SeDJ-1 stimulation of PI3K/AKT pathway. In contrast, SeH4 functions as an inhibitor of asexual and sexual development.
Materials and methods
Strains and culture conditions
We generated or used the following strains in this study to explore the function of Seasf1: Stemphylium eturmiunum strain (EGS 29-099) was cultured on PDA or CM medium (casein acid hydrolysate 0.5 g/L, casein enzymatic hydrolysate 0.5 g/L, glucose 10.0 g/L, Ca(NO3)2·4H2O 1.0 g/L, KH2PO4 0.2 g/L, MgSO4·7H2O 0.25 g/L, NaCl 0.15 g/L, yeast extract 1.0 g/L, and agar 15.0 g/L) at 25 °C under the dark condition. Escherichia coli DH5α and Agrobacterium tumefaciens AGL-1were incubated in LB (Luria–Bertani) medium at 37 °C or 28 °C (Lennox 1955).
Generation of asf1 knockout mutants in S. eturmiunum
Cloning and propagation of recombinant plasmids (Table S1) were done under standard conditions (Sambrook and Russell 2001). The knockout vector pXEH carrying an Hph resistance cassette was used in this study. Seasf1 was cloned from the S. eturmiunum strain upon its genome sequence (did not upload). Seasf1-pXEH vector was constructed as follows: the flanking regions of Seasf1, including 1500 bp upstream and 1500 bp downstream of its open reading frame, were separately amplified using primer pairs (Seasf1-5f/Seasf1-5r and Seasf1-3f/Seasf1-3r) (Table S2). The upstream fragment of Seasf1 was inserted into pXEH vector by XhoI/BglII digestion. The downstream fragment of Seasf1 was then inserted into pXEH vector via BamHI/XbaI digestion. To obtain Seasf1 knockout mutants, Seasf1-pXEH vector was transformed into S. eturmiunum strain using Agrobacterium tumefaciens-mediated transformation (ATMT) (Bernardi-Wenzel et al. 2016). The knockout strains were screened on HygB-resistance by PCR and southern blot. Then, nine other genes, including Sedj-1, SeH4, Sepi3k, Se01950, Se03485, Se04320, Se07693, Se10206 and Se10302, were deleted upon the same method. The primers for obtaining the mutants of these nine genes were shown in Table S2.
Generation of Seasf1 complement transformants
To generate complement transformants of Seasf1, Seasf1 was inserted into EGFP-pHDT vector. The EGFP-pHDT-Seasf1 was obtained and then transformed into ∆Smasf1 (Sm: Sordaria macrospora) (S90177) and Seasf1 knockout strains, respectively. These experiments were carried out by ATMT method (Bernardi-Wenzel et al. 2016). All these transformants were screened by G418-resistantand further confirmed by PCR and western blot.
Generation of the tested genes overexpression transformants
To generate Sedj-1 overexpression transformants, Sedj-1 was cloned from S. eturmiunum strain using the primers (Table S2) and then inserted into the EGFP-pHDT vector to create recombinant plasmids EGFP-pHDT-Sedj-1. The recombinant plasmids were transformed into the Sedj-1 knockout mutants using the ATMT method (Bernardi-Wenzel et al. 2016). Overexpression transformants resistant to G418 were screened using qRT-PCR and western blot. Overexpression transformants of Sepi3k or truncations of Sedj-1 were produced by the same method.
Southern blot
Seasf1 knockout and S. eturmiunum strains were inoculated in PDA medium and grown at 25 °C for 7 days in the dark condition. Genomic DNA was extracted from mycelia of Seasf1 knockout or S. eturmiunum strains by CTAB (Štorchová et al. 2000). Southern blot was performed using the DIG High Prime DNA Labeling and Detection Starter kit (version I) upon the manufacturer’s instructions (Roche Diagnostics, Mannheim, Germany). The Hph gene was amplified from S. eturmiunum strain using primer pairs (Table S2) and produced a DIG-labeled probe for hybridization. Each experiment was repeated at least three times.
RNA extraction and qRT-PCR
The mycelia of all tested strains, including overexpression transformants and S. eturmiunum stains, were inoculated in PDB (Potato Dextrose Broth) cultures at 25 °C for 4 days. For RNA extraction, the mycelia were frozen and powered in liquid nitrogen followed by extraction using the Fungal RNA Kit (OMEGA Biotechnology, USA). Reverse transcription was done using 1 µg of total RNA per 20 µL reaction. SYBR Color qRT-PCR was performed in 20 µL reactions system, including 0.4 µg of cDNA, 0.4 µL of gene-specific primers (Table S2), 10 µL of 2× SYBR Green qPCR Mix (Shandong Sparkjade Biotechnology Co., Ltd.), and 5.2 µL of ddH2O. The qRT-PCR was performed on an ABI QuantStudio™ 6 Quantitative Real-Time PCR System (Applied Biosystems) under the following conditions: 95 °C for 5 min, 40 cycles at 95 °C for 10 s, and 60 °C for 30 s to calculate cycle threshold values, followed by dissociation at 95 °C for 15 s, 60 °C for 1 min, and 95 °C for 15 s to obtain melting curves. Relative expression levels of selected genes were calculated using the 2−∆∆CT method (Livak and Schmittgen 2001). The housekeeping gene actin was used as an internal standard in each case. This experiment was repeated technically three times.
RNA sequence analysis
For total RNA extraction, the mycelia of Seasf1 knockout mutants and S. eturmiunum strains were grown for 4 days in PDB medium, and RNA was then extracted using the Fungal RNA Kit (OMEGA Biotechnology, USA). The eligible mRNA was enriched using magnetic beads with Oligos (dT), and then broken into short fragments using Fragmentation buffer. The mRNA of Seasf1 knockout mutants or S. eturmiunum was used as a template. First-strand cDNA was synthesized with random hexamers. Second-strand cDNA was synthesized by adding buffer, dNTPs, and DNA polymerase I to the mixture of first-strand cDNA. AMPure XP beads were used to purify the double-stranded cDNA. Subsequently, the purified double-stranded cDNA was subjected to end repair, annealed to sequencing adapters, and selected fragment size using AMPure XP beads. Finally, PCR enrichment was performed to obtain a final cDNA library.
RNA sequencing (RNA-Seq) analysis was performed on cDNA library from the Seasf1 knockout mutants or S. eturmiunum stains using Illumina Hiseq4000 sequencing (Berry Genomics, Beijing). Approximately 300 bp fragments were inserted into every library, in which 100 bp sequences were read. Low-quality raw reads were filtered out. Resulting paired-end sequencing reads were aligned and quantified using TopHat and Cufflinks with default parameter values. Denovo transcriptome analysis was used to estimate transcript abundance and differential expression. Gene expression was calculated as fragments per kilobase of transcript per million mapped fragments (FPKM).
Gene ontology
The data of transcriptomic analysis was used the Gene Ontology (GO) Biological Process, KEGG (Kyoto Encyclopedia of Genes and Genomes), Swiss-Prot (a manually annotated and reviewed protein sequence database), PIR (Protein Information Resource), and PRF (Protein Research Foundation) databases at FungiDB (Stajich et al. 2012) and Blast2GO v2.5. To characterize the differentially expressed genes (DEGs), GO-based trend tests were carried out using Fisher’s exact test. A fold change greater than 2.0 and a P value less than 0.05 were considered statistically significant.
Phosphopeptide enrichment and Nano-UHPLC-MS/MS analysis
Hyphae of both Seasf1 knockout mutants and S. eturmiunum strains were collected into an Eppendorf tube, and then milled immediately with lysis buffer (1% SDS, 8 M urea, and 1× Protease Inhibitor Cocktail (Roche Ltd. Basel, Switzerland) and vibrated three times for 5 min. The sample was then lysed on ice for 30 min and centrifuged at 15,000 rpm for 15 min at 4 °C. The supernatant was collected and transferred into a new Eppendorf tube. The protein concentration was determined by Bicinchoninic Acid (BCA) protein assay. Approximately 1 mg protein was then transferred into a new Eppendorf tube and adjusted to a volume of 400 μL with 8 M urea, followed by adding 20 μL of 0.5 M Tris (2-carboxyethyl) phosphine (TCEP). After incubating at 37 °C for 1 h, 40 μL of 1 M iodoacetamide was added into the sample to incubate for 40 min under the dark condition at 25 °C.
Five volumes of pre-chilled acetone were poured into the samples to precipitate the protein overnight at − 20 °C. The samples were centrifuged for 20 min at 14,000 rpm (4 °C). The precipitates were washed twice using 1 mL pre-chilled 90% acetone aqueous solution and then dissolved in 400 μL of 100 mM Tetraethylammonium Bromide (TEAB). Trypsin (Promega, Madison, WI) was added into the mixture of TEAB at the ratio of 1:50 (enzyme: protein, mass ratio) to digest the proteins overnight at 37 °C. For phosphopeptide enrichment, the digested peptide mixture was desalted by C18 ZipTip and quantified by Pierce™ Quantitative Colorimetric Peptide Assay (23275). The sample was selectively enriched with High-Select™ TiO2 Phosphopeptide Enrichment Kit (Thermo Fisher Scientific, MA, USA) following the manufacturer’s instructions, and dried immediately in a speed vacuum concentrator. These peptides were re-dissolved in 0.1% FA (formic acid) and analyzed by nanospray UHPLC-MS/MS.
Yeast two-hybrid
For yeast two-hybrid analysis, Y2H assay was performed by the Yeast Protocols Handbook (Clontech) using the Y2H Gold yeast reporter strain (Clontech). SeH3, SeH4, Sepi3k, Segsk3, and truncations of SeDJ-1-M1, SeDJ-1-M2, SeDJ-1-M3, SeDJ-1-M4, SeDJ-1-M5, SeDJ-1-M6, and SeDJ-1-M7 were amplified from S. eturmiunum strain using PCR (Table S2). The PCR products were purified and digested with restriction enzymes. Seasf1, SeH4, Sedj-1and Sepi3k were inserted separately into pGBKT7 plasmids (BD). SeH4, SeH3, Sedj-1, Sepi3k, Segsk3, and truncations of Sedj-1 were inserted separately into pGADT7 plasmids (AD). The recombinants of Seasf1-BD and SeH4-AD, Seasf1-BD and SeH3-AD, Seasf1-BD and Sedj-1-AD, SeH4-BD and Sedj-1-AD, Sedj-1-BD and Sepi3k-AD, Sedj-1-BD and Segsk3-AD, and Sepi3k-BD and SeDJ-1-M-AD (SeDJ-1-M1, SeDJ-1-M2, SeDJ-1-M3, SeDJ-1-M4, SeDJ-1-M5, SeDJ-1-M6 and SeDJ-1-M7) were co-transformed into the Y2H gold yeast strain. The co-transformants were screened on SD/-Trp/-Leu medium (TaKaRa Bio) at 30 °C for 3–5 days and assayed for growth on SD/-Trp/-Leu/-His/-Ade/X-α-gal plates (TaKaRa Bio). Each experiment was repeated at least three times.
Pull-down
The Seasf1, Sepi3k or Segsk3 was cloned from S. eturmiunum strain into pET28a vector after adding a 1 × FLAG tag to the 5′-terminal of their by PCR (Table S2). Seasf1 or Sedj-1 was inserted into pGEX-6P-1 vector. For expression of Flag-SeASF1-28a, Flag-SePI3K-28a, Flag-SeGSK3-28a, GST-SeASF1 and GST-SeDJ-1, constructs of the pET28a or pGEX-6P-1 was transformed separately into E. coli Transetta (DE3) (Transgene, Beijing, China). These cells were grown to OD600 = 0.6–0.8 at 37 °C, induced with 1 M Isopropyl-β-D-Thiogalactoside (IPTG) for 12–16 h at 16 °C, and harvested by centrifugation for 5 min using 8000 rpm at 4 °C. The Flag-SeASF1-28a cells were resuspended in Ni-lysis buffer (30 mM Tris–HCl, 300 mM NaCl, 30 mM Imidazole, pH 7.5) and lysed with an Ultrasonic Cell Disruptor. The lysate was centrifuged for 30 min at 14,000 rpm (4 °C), and the supernatant was passed over a Ni-affinity column (GE) at least three times. The protein of Flag-SeASF1-28a was eluted using Ni-elution buffer (30 mM Tris–HCl, 300 mM NaCl, 6 M Imidazole, pH 7.5). Flag-SePI3K-28a or Flag-SeGSK3-28a cells were resuspended in Ni-lysis buffer (50 mM Tris–HCl, 500 mM NaCl, 30 mM Imidazole, pH 8.0) and lysed with an Ultrasonic Cell Disruptor. The lysate was centrifuged for 30 min at 14,000 rpm (4 °C), and the supernatant was passed over a Ni-affinity column (GE) at least three times. The protein of Flag-SePI3K-28a or Flag-SeGSK3-28a was eluted using Ni-elution buffer (50 mM Tris–HCl, 500 mM NaCl, 6 M Imidazole, pH 8.0). The GST-SeASF1 or GST-SeDJ-1 cells were resuspended in GST-lysis buffer (50 Mm HEPES, 500 mM NaCl, pH 8.0) and lysed with an Ultrasonic Cell Disruptor. The lysate was centrifuged for 30 min at 14,000 rpm (4 °C), and the supernatant was passed over a GST-affinity column (glutathione sepharose™ 4B beads GE Healthcare, Little Chalfont, Buckinghamshire, UK) at least three times. The protein of GST-SeASF1 or GST-SeDJ-1was eluted by GST-elution buffer (50 mM HEPES, 500 mM NaCl, 10 mM L-glutathione, pH 8.0). The eluent proteins were mixed with loading buffer and validated using SDS-PAGE.
For pull-down, GST-SeDJ-1 and Flag-SeASF1-28a were expressed respectively in E. coli strain BL21 (DE3) (Transgene, Beijing, China). Total proteins from the GST-SeDJ-1 and Flag-SeASF1-28a strains were then incubated with 4000 μL of glutathione sepharose™ 4B beads for 2 h at 4 °C. The supernatant was removed and the beads were washed with GST-lysis buffer three times. Finally, the beads were eluted with GST-elution buffer. Pull-down of GST-SeDJ-1 with Flag-SeASF1-28a was detected using an anti-Flag (Abclonal, AE005). Each experiment was repeated at least three times. The pull-down of GST-SeDJ-1 and Flag-SePI3K-28a, or GST-SeDJ-1 and Flag-SeGSK3-28a was performed using the same approaches.
Co-IP
For Co-IP assay, Seasf1, SeH4, Sedj-1, Sedj-1-M6, and Sedj-1-M7 were inserted into pDL2 vector, respectively. SeH3, SeH4, Sedj-1, Sepi3k, and Segsk3 were inserted into pFL7 vector alone. Each of these constructs was done in yeast (XK125) using a recombination approach (Zhou et al. 2011). Recombinant plasmids were then co-transformed into the protoplasts of Fusarium graminearum wild-type strain (PH-1). Transformants were detected using western blot. For FLAG IP, total proteins of transformants were extracted with an extraction buffer [50 mM HEPES, 130 mM NaCl, 10% glycerin, protease inhibitors (25 mM Glycerol phosphate, 1 mM Sodium orthovanada, 100 mM PMSF), pH 7.4]. Subsequently, protein extracts were incubated with 30 μL of Affinity Gel-conjugated Mouse anti DDDDK-Tag mAb (ABclonal Technology (WuHan, China), AE061) for 4 h at 4 °C. The beads were collected by centrifugation at 3000 rpm and washed five times with a washing buffer (50 mM HEPES, 130 mM NaCl, 10% glycerin, pH 7.4). The bound proteins were eluted from the beads by boiling them for 15 min. The beads were collected using centrifugation for 2 min at 3000 rpm. Proteins were separated on 12% SDS–PAGE gels and detected using immunoblot with a monoclonal α-Flag antibody (Abclonal, AE005) or α-GFP antibody (Abclonal, AE012). Membranes were stained with Ponceau solution (CWBIO, Beijing, China). Each experiment was repeated at least three times.
Western blot
Protein samples were separated on 12% SDS-PAGE gels at 100 V for 3 h in running buffer (25 mM Tris-base, 200 mM Glycine, 0.1% SDS). Gels were transferred to an Immobilon®-P PVDF membrane for 1.5 h at 230 mA. Membranes were blocked in 5% non-fat milk in 1 × TBST (0.02 M Tris-base, 0.14 M NaCl, pH 7.4) with 0.1% (vol/vol) Tween-20 prior to addition of GFP (Abclonal, AE012) or Flag antibodies (Abclonal, AE005) at 1:5000 dilution and incubated at room temperature for 1–1.5 h. The membranes were washed three times with TBST (Tris-base Buffered Saline Tween) and incubated for 1 h with a horseradish peroxidase-labeled immunoglobulin G (IgG-HRP) secondary antibody-Goat Anti-Mouse IgG (H + L) HRP (ABclonal) at 1:7500 dilution. The specific proteins were visualized using an ECL Chemiluminescence Detection Kit (Vazyme Biotech Co., Ltd, Nanjing, China). Images were taken using a Tanon-5200 Chemiluminescent Imaging System (Tanon, Shanghai, China). Each experiment was repeated at least three times.
Microscopy
To observe the morphology of conidia and conidiophores, all the transformants and WT strains were grown side-by-side in the dark at 25 °C for 4 weeks on CM medium by inserting double slides. To examine the distribution of nuclear in mycelia, the transformants and WT strains stained using the 4, 6-diamidino-2-phenylindole (DAPI). To image sexual structures such as pseudothecia, asci, and ascospores, all tested strains were cultured on PDA medium at 25 °C for 6 weeks under the dark condition. Pseudothecia were sectioned using a double-edged blade under a dissecting microscope (Olympus, SZX10). The asci, conidia and conidiophores were captured with 20× or 40× objective lenses on an Olympus microscope (Olympus BX53, Tokyo, Japan) using differential interference contrast (DIC) and fluorescence illumination. The experiment was repeated at least three times.
Statistical analysis
Data were analyzed using Systat 12 (Systat Software Inc., San Jose, CA, USA). The data were subjected to the one-way analysis of variance (ANOVA) test. A two-way Student’s t test and Duncan’s multiple range test of least significant difference (LSD) were used when there were more than two means. P values lower than 0.05 and 0.01 were considered statistically significant.
Results
SeASF1 regulates sexual reproduction in Sordaria macrospora
ASF1 was identified from the Stemphylium eturmiunum genome database (unpublished). This gene was named SeASF1 (KX033515). SeASF1 has 291 amino acids with a calculated molecular mass of 31.98 kDa. Alignment of the SeASF1 sequence with its homologous sequences from plants, animals, and other fungal species (https://www.ncbi.nlm.nih.gov/) (Table S3) revealed that the N-terminus sequences (1–154 aa) of ASF1 was highly conserved and contained the ASF1 hist chap superfamily functional domain, whereas the C-terminus was varied greatly between species (Fig. S1). Phylogenetical analysis grouped these genes into three clusters corresponding to fungi, animals, and plants. The fungi group could further be subdivided into clusters which corresponded with taxonomy. SeASF1 shares 98.28% similarity with ASF1 from S. lycopersici and 90.51–91.13% with ASF1 from two Pyrenophora species. Also, SeASF1 shares 92.78% identity with ASF1 from Setosphaeria turcica and more than 91% with ASF1 from five Bipolaris species. However, SeASF1 shares 45.45% with ASF1 from Schizosaccharomyces pombe (Fig. S2). All these data indicate that ASF1 is widely distributed in a number of fungi species. In a previous study, the S. macrospora ASF1 (XP003345657) was localized to the nucleus and was essential for sexual reproduction (Gesing et al. 2012). In Fig. S2, all the ASF1 sequences share a conserved functional domain, thus, we hypothesize that Seasf1 plays a similar role in sexual development in most fungi species. When Seasf1 was heterologously expressed in the ∆Smasf1 strain, ∆Smasf1::EGFPSeasf1-1 and ∆Smasf1::EGFPSeasf1-2, complemented the sexual defects of the ∆Smasf1 strains (Fig. 1a, b). PCR and western blot were used to identify the genetic constructs (Fig. S3). In contrast to ∆Smasf1 strain, the nuclear distributions in two heterologous transformants were in accord with those of in WT::EGFP and WT strains (Fig. 1c). Together, SeASF1, similar to SmASF1, plays an important role in regulating sexual reproduction.
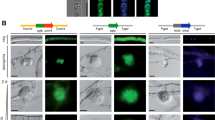
Seasf1 contributes to the sexual development in Sordaria macrospora. To identify whether Seasf1 could influence the sexual characterization of S. macrospora, we obtained two Seasf1 complement transformants through its transformed into ∆Smasf1 of S. macrospora. a Perithecia of four transformants and WT strains were visualized as black structures on PDA medium. ∆Smasf1 was Smasf1 deleted mutants in S. macrospora. ∆Smasf1::GFPSeasf1-1/2 were obtained by Seasf1 heterologous expression in ∆Smasf1. WT::GFP: GFP was transformed into WT strain. WT was S. macrospora wild type strain. All strains were cultured on PDA medium for 10 days, respectively. WT::GFP and WT were used as controls. The small boxes on the bottom right sides were enlarged perithecia. b Asci and ascospores of ∆Smasf1::GFPSeasf1-1/2, WT::GFP and WT strains were produced on PDA medium for 20 days. c Nuclear distribution in four transformants and WT strains were observed by fluorescence microscopic. Bar = 10 μm, 20 μm, 50 μm and 500 μm
SeASF1 modulates asexual and sexual development in S. eturmiunum that is not correlated with MAPK signaling pathway
To study the biological functions of Seasf1 during vegetative and sexual development in S. eturmiunum strain, we produced two Seasf1 knockout mutants (∆Seasf1-0::EGFP and ∆Seasf1-5::EGFP) and two complement transformants (∆Seasf1-0::EGFPSeasf1 and ∆Seasf1-5::EGFPSeasf1). These four transformants were identified using PCR, western blot, and southern blot, respectively (Figs. S4, S5). To study how Seasf1 affected growth, these four transformants and WT strains were inoculated on PDA medium and photographed after 1, 3, 5, 7 and 9 days (Fig. S6a). In comparison to WT strain, the colony diameters of two ∆Seasf1 mutants were more smaller than two complement transformants and WT strains, but two complement transformants almost had normal colonies growth as well as WT strain (Fig. S6a–c). In contrast to complement transformants and WT strains, two ∆Seasf1 mutants exhibited hyphal fusion, anomalous distribution of nuclei in the mycelium and increased number of malformed conidia (Fig. 2a–e). The two ∆Seasf1 mutants did not produce pseudothecia, asci and ascospores in contrast to the two complement transformants and WT strains (Fig. 2f, g). These results suggest that Seasf1 is involved in asexual and sexual development in S. eturmiunum.
The asexual and sexual development was regulated by SeASF1 in S. eturmiunum. The asexual and sexual characteristics were observed between Seasf1 knockout mutants and complement transformants compared with WT strain. Two ∆Seasf1 mutants were obtained by deleting Seasf1 in S. eturmiunum. Two ∆Seasf1::EGFPSeasf1 transformants were produced by Seasf1 expression in ∆Seasf1. WT was S. eturmiunum wild type strain. a Hyphal fusion showed in four transformants and WT strains, respectively. The images were photographed after growing on PDA medium for 7 days. The fusions in the hyphae were marked with white arrows. b The mycelia of four transformants and WT strains were examined by DIC and fluorescence microscopy for GFP and DAPI after growing on PDA medium for 12 days. c Conidia morphologies of four transformants and WT strains were cultured on CM medium for 4 weeks. d The numbers of conidia were counted by blood counting chamber. e Percentages of deformed conidia were analyzed in two ∆Seasf1 mutants compared with two ∆Seasf1::EGFPSeasf1 transformants and WT strains. f Pseudothecia of four transformants and WT strains were visualized as black structures on PDA medium. Pseudothecia were shown in the lower right. g Asci and ascospores of four transformants and WT strains were merged using microscope after cultured on PDA medium for 6 weeks. Bar = 20 μm and 500 μm
The mitogen-activated protein kinase (MAPK) signaling pathway was a key regulator of asexual and sexual development in fungi species (Bardwell et al. 1996; Errede et al. 1993; Izumitsu et al. 2009; Maerz et al. 2008; Neiman and Herskowitz 1999; Teichert et al. 2014). Five genes Gα subunit, Mat1, Mat2, Ste2 and Ste3 were essential for fungi sexual development (Lin et al. 2005, 2011; Liu and Dean 1997; Liu et al. 2018; Metin et al. 2010; Vitale et al. 2019) and were major components of the MAPK pathway (Crespo et al. 1994; Herskowitz 1995; Müller et al. 1999; van Drogen et al. 2001; Whiteway et al. 1995). To determine whether these five genes contributed to how ASF1 regulated asexual and sexual development in S. eturmiunum, we measured the expression levels of these genes in two ∆Seasf1 mutants relative to WT strains. However, we noted that the expression levels of these genes did not change significantly in two ∆Seasf1 mutants (Fig. S7). Asexual and sexual development by the MAPK pathway can be regulated by protein phosphorylation (Kawasaki et al. 2002; Millar et al. 1995). To determine whether any of these five genes were phosphorylated in response to expression of SeASF1, a quantitative proteomics assay was implemented in ∆Seasf1 and WT (Fig. S8). Thirty proteins were phosphorylated in MAPK signaling pathway, but this list did not include any of these proteins (Table S4). Taken together, these data suggest that these five genes are not involved in ASF1 mediation of S. eturmiunum asexual and sexual development, thus, ASF1 is impossible to trigger MAPK signaling pathway in S. eturmiunum. Therefore, other an unknown signaling pathway is likely to mediate SeASF1 to modulate asexual and sexual development.
SeDJ-1 mediates SeASF1 to regulate asexual and sexual development of S. eturmiunum
To investigate whether other genes involved in the functions of SeASF1, we analyze the data of ∆Seasf1 transcriptome compared with that of the WT-vegetative and WT-sexual phases. These results show that 2342 genes were up-regulated and 1374 genes were down-regulated between ∆Seasf1 and WT-sexual; 1719 were up-regulated and 1304 were down-regulated between ∆Seasf1 and WT-vegetative; also, 1343 were up-regulated and 1751 were down-regulated between WT-sexual and WT-vegetative (Fig. S9b). A total of 380 differentially expressed genes (DEGs) were identified among three transcriptomes (fold change > 2.0, p value < 0.05) and subsequently analyzed using hierarchical clustering and comparative analysis (Figs. S9a, S10). We speculated that these significantly up-regulated or down-regulated genes might be involved in SeASF1-regulated asexual and sexual development.
GO enrichment showed that these DEGs had functions related to cellular processes secondary metabolite development, organelle activity and catalytic activity (Fig. S9c, d). Seven genes, including SeDJ-1 (transcription factor), Se01950 (Heat shock protein), Se03485 (LysM domain-containing protein), Se04320 (Proline dehydrogenase), Se07693 (vesicle coat complex COPII, subunit SEC31), Se10206 (Allantoate permease), and Se10302 (Choline dehydrogenase), were considered as potential interactors with SeASF1 (Table S5). However, Y2H analysis showed that only SeDJ-1 interacted directly with SeASF1 (Fig. S11). Phylogenetic analysis showed that DJ-1 was widely distributed in plants, animals, and other fungal species (Table S6) (Fig. S12). SeDJ-1 shared 93.77% sequence similarity with DJ-1 from S. lycopersici (RAR14805) but less than 20% similarity with DJ-1 from all other fungal species. Thus, SeDJ-1 is possible to play important roles in regulating asexual and sexual development in both S. eturmiunum and S. lycopersici.
To investigate whether these seven genes affected asexual and sexual development in S. eturmiunum, two knockout mutants were generated for each gene by homologous recombination and mutants were inoculated onto agar plates alongside the WT strain. As a result, the mutations of SeDJ-1 inhibit the rate of asexual and sexual development (Figs. S14–S19). Two Sedj-1 knockout mutants (∆Sedj-1–1 and ∆Sedj-1–4) appeared to slow the growth rate of colonies relative to the WT strain (Fig. S13a, b). In contrast to WT strain, the asexual and sexual development of ∆Sedj-1–1 and ∆Sedj-1–4 were significantly altered, and the nuclei showed anomalously distribution in the mycelia (Fig. 3a). Conidiogenous cells in the two mutants were swollen either at the apex or the lateral branch, whereas they grew to be secondary mycelia at the apex or on the side of WT strain within 7 days. By day 13, the conidiophores and conidiogenous cells in the two mutants imaged the pale, whereas conidiogenous cells in WT strain appeared swollen at the apex and darkened. In addition, conidia bodies were young, solitary, brown, and ellipsoid to cylindrical in shape. By day 20, subglobose and young conidia were found in two mutants, whereas WT strain produced nearly mature, oblong conidia. By day 30, two mutants had not developed mature conidia and their conidiophores became abnormally bead-like (Fig. 3b). By day 13, young, irregular ascogonia produced in the two mutants, whereas the young protoperithecia had developed in the WT strain. By day 25, pseudothecia did not produce in the two mutants, opposing to WT strain. At 34 days, two mutants did not produce mature pseudothecia, while the WT strain had produced nearly mature pseudothecia with asci. Lastly, mature asci were only found in WT strain by day 45 (Fig. 3c). These results indicate that SeDJ-1 plays an essential role in the asexual and sexual development in S. eturmiunum.
The characterization was indicated the role of SeDJ-1 in asexual and sexual development of S. eturmiunum. The asexual and sexual characteristics were observed in Sedj-1 knockout mutants (∆Sedj-1-1 and ∆Sedj-1-4) compared with WT strain. Two mutants were obtained by deleting Sedj-1 in S. eturmiunum. Two mutants and WT strains were cultured on PDA or CM medium, respectively. WT was S. eturmiunum (EGS 29-099) strain. a The mycelium of two mutants and WT strains were incubated on PDA medium for 6 days and examined by DIC and fluorescence microscopy. The nuclei of the mycelia were discovered under the fluorescence microscopy after staining by DAPI. b For the microscopic observation of conidiophores, conidiogenous cells and conidia development, two mutants and WT strains were grown on CM medium for 7 days, 13 days, 20 days, and 30 days respectively. Black arrowheads indicated conidiogenous cells, and blue arrowheads indicated secondary mycelia. c For the microscopic observation of ascogonia, protoperithecia, young pseudothecia and asci development, all strains were grown on PDA medium and examined after growth at 25 °C for 13 days, 25 days, 34 days, and 45 days respectively. Insets showed enlarged ascogonia and protoperithecia on the bottom right sides. Pseudothecia were visualized as black structures. Black arrows indicated ascogonia, and red arrows indicated protoperithecia. Bar = 20 μm, 50 μm and 500 μm
To further investigate whether interactions occur among SeASF1, SeDJ-1, SeH3 and SeH4, we used Y2H, GST pull-down and Co-IP to evaluate the interactions among them. As a result, SeASF1 binds to SeH4 and SeDJ-1 but does not with SeH3, while SeDJ-1 interacts with SeH4 (Fig. 4). Next, qRT-PCR was used to monitor the transcript levels of Sedj-1 and SeH4 in two ∆Seasf1 mutants, and the expression levels of Seasf1 and SeH4 in two ∆Sedj-1 mutants. The expression levels of Sedj-1 and SeH4 showed down- and up-regulation in two ∆Seasf1 mutants respectively (Fig. S20a). In two ∆Sedj-1 mutants, the expression levels of SeH4 showed down-regulation, but those of Seasf1 did not alter (Fig. S20c). We speculate that SeDJ-1 or SeH4 combinates with SeASF1 to modulate asexual and sexual development in S. eturmiunum, but ∆Sedj-1 does not alter the expression of SeASF1. Thus, SeASF1 is not possible to involve in SeDJ-1 regulation of asexual and sexual development in S. eturmiunum.
Interactions among SeASF1, SeDJ-1, SeH3, and SeH4 in S. eturmiunum. The interactions of SeASF1, SeDJ-1, SeH3 and SeH4 were verified using Y2H, GST pull-down, and Co-IP assays. a SeASF1 was cloned into the bait plasmid pGBKT7 (BD). SeH4 or SeH3 was cloned into the prey plasmid pGADT7 (AD). Yeast transformants were first grown on SD/-Trp/-Leu (DDO), and then selected on SD/-Trp/-Leu/-His/-Ade/X-α-gal (QDO/X) for activating X-a-galactosidase activity. b SeDJ-1 was cloned into the prey plasmid pGADT7 (AD). SeH4 was cloned into the bait plasmid pGBKT7 (BD). The images were photographed at 3 days after incubation. Each experiment was repeated at least three times. c SeASF1 was cloned into plasmid pGEX-6P-1. SeH4 or SeH3 was cloned into plasmid pET28a. SeASF1-GST was expressed in E. coli and incubated with SeH4-His or SeH3-His, purified (pull-down) by glutathione sepharose beads. Recombinant GST was used as control. SeH4-His was pulled down by SeASF1-GST. d SeASF1 was cloned into plasmid pDL2, SeH4 or SeH3 was cloned into plasmid pFL7. Total proteins were extracted from F. graminearum protoplasts expressing SeASF1-GFP, SeH4-Flag, and SeH3-Flag. Recombinant GFP or Flag was used as control. The immune complexes were immunoprecipitated with α-Flag antibody (α-Flag IP). Coprecipitation of SeH4-Flag or SeH3-Flag was detected by immunoblotting. e SeDJ-1 was cloned into plasmid pGEX-6P-1. Flag-SeASF1 or SeH4 was cloned into plasmid pET28a. SeH4-His and Flag-SeASF1-His were retained by SeDJ-1-GST respectively. f SeH4 was cloned into plasmid pDL2. SeDJ-1 was cloned into plasmid pFL7. Total proteins were extracted from F. graminearum protoplasts expressing SeASF1-GFP, SeH4-GFP, and SeDJ-1-Flag respectively. Coprecipitation of SeDJ-1-Flag was detected by immunoblotting. Membranes were stained with Ponceau S to confirm equal loading. Protein sizes were indicated in kDa. Each experiment was repeated at least three times
To address the role of SeH4 in S. eturmiunum. We obtained two ∆SeH4 mutants (∆SeH4-8 and ∆SeH4-20) using homologous recombination, but these two mutants did not manipulate asexual and sexual development of S. eturmiunum (The data not shown). The expression of Sedj-1 or Seasf1 was almost unchanged in two ∆SeH4 mutants (Fig. S20b). Thus, SeH4 does not affect asexual and sexual development, and SeH4 interacts with SeASF1 and SeDJ-1, indicating SeH4 may be an auxiliary factor to involved in SeASF1-regulated asexual and sexual development, but not a necessary component in this pathway. Then, two Sedj-1 overexpression transformants, OESedj-1::∆Seasf1-4 and OESedj-1::∆Seasf1-6, were obtained by transformation Sedj-1 into the ∆Seasf1 mutants using ATMT method, as well as, both OESeasf1::∆Sedj-1–3 and OESeasf1::∆Sedj-1–8 were obtained by transformation Seasf1 into the ∆Sedj-1 mutants. Both OESedj-1::∆Seasf1-4 and OESedj-1::∆Seasf1-6 complemented the asexual and sexual phenotype of ∆Seasf1 but OESeasf1::∆Sedj-1–3 and OESeasf1::∆Sedj-1–8 did not complement the mutant phenotype of ∆Sedj-1 (Fig. S21). Taken together, SeDJ-1 can operate SeASF1 to regulate asexual and sexual development of S. eturmiunum but SeASF1 is not required for the functions of SeDJ-1.
SeDJ-1 is required for SePI3K to modulate asexual and sexual development of S. eturmiunum
The PI3K/AKT signaling pathway manipulates a variety of biological processes, but the role of this pathway in fungi development has not been addressed. In animals, DJ-1 activated the PI3K/AKT signaling pathway (Yang et al. 2005; Zhang et al. 2016) via PI3K and GSK3 (Oh and Mouradian 2017), while the functions of this pathway would be significantly altered when DJ-1 perceived these two crucial components (Lee et al. 2021; Sitaram et al. 2009; Wang et al. 2013). To determine whether this pathway was conserved in fungi, the expression levels of Sepi3k and Segsk3 were quantified in two ∆Sedj-1 mutants and two ∆Sepi3k mutants were obtained by deleting Sepi3k in S. eturmiunum, and the expression levels of Sedj-1 and Segsk3 were measured in these two mutants. The expression levels of Sepi3k or Sedj-1 were down-regulated in two ∆Sedj-1 or two ∆Sepi3k mutants, while the expression levels of Segsk3 were up-regulated in these four mutants (Fig. S22a, b). Our experiments indicated that Sedj-1 was likely to regulate positively Sepi3k to effect asexual and sexual development in S. eturmiunum. Inversely, Segsk3 was a negative regulator of Sedj-1 and Sepi3k, but it was not able to influence the asexual and sexual development of S. eturmiunum. These experiments suggested that SeDJ-1 mediated these affects via the PI3K/AKT signaling pathway in S. eturmiunum. We used Y2H, Co-IP, and Pull down to demonstrate that SeDJ-1 simultaneously interacted with SePI3K and SeGSK3 (Fig. S23). Together, these results suggests that the developmental activator, SeDJ-1, mediates the asexual and sexual development of S. eturmiunum via SePI3K or PI3K/AKT signaling pathway.
Next, we showed by truncation mutation analysis and Y2H and Co-IP that the segment SeDJ-1-M6 which consisted of amino acids 134–175 was responsible for binding with SePI3K (Fig. S24a, b). Subsequently, Sedj-1, Sepi3k, Sedj-1-M6, and Sedj-1-M7 were overexpressed in ∆Sepi3k mutants alone. Eight transformants, such as OESedj-1-T13 and OESedj-1-T30, OESedj-1-M6-T5 and OESedj-1-M6-T10, OESedj-1-M7-T18 and OESedj-1-M7-T26, and OESepi3k-T8 and OESepi3k-T13, were then obtained upon qRT-PCR and western blots (Fig. S25a, b). All these transformants except for OESedj-1-M7-T18 and OESedj-1-M7-T26 restored asexual and sexual characteristics compared with two ∆Sepi3k mutants (Fig. 5a, b). Otherwise, the nuclei distributions in mycelia of these transformants except for OESedj-1-M7-T18 and OESedj-1-M7-T26 were almost consistent with WT strain in contrast to two ∆Sepi3k mutants (Fig. 5c). Four overexpression transformants, OESepi3k-T8 and OESepi3k-T12, and OESedj-1-T5 and OESedj-1-T20, were produced by overexpression of Sepi3k or Sedj-1 in ∆Sedj-1 mutants, and then verified by qRT-PCR and western blots (Fig. S26a, b). In contrast to ∆Sedj-1 mutants, these four transformants recovered asexual and sexual characteristics relative to WT (Fig. S26c, d). The nuclei distributions in mycelia of these four transformants appeared to the same patterns as WT, opposing to two ∆Sedj-1 mutants (Fig. S26e). All these results indicated that SeDJ-1, especial SeDJ-1-M6 segment, carried out the same roles as the SePI3K in S. eturmiunum. Significantly, SePI3K is a pivotal component in the PI3K/AKT signaling pathway in S. eturmiunum, thus, SeDJ-1, as well as SePI3K, can activate the PI3K/AKT signaling pathway to modulate asexual and sexual features in S. eturmiunum.
Sedj-1 or Sedj-1-M6 recovers ∆Sepi3k mutants to regulate the asexual and sexual characteristics in S. eturmiunum. The asexual and sexual characteristics were observed in eight overexpression transformants. Eight overexpression transformants, including OESedj-1-T13 and OESedj-1-T30, OESedj-1-M6-T5 and OESedj-1-M6-T10, OESedj-1-M7-T18 and OESedj-1-M7-T26, and OESepi3k-T8 and OESepi3k-T13, were obtained by overexpressing Sedj-1, Sepi3k, Sedj-1-M6, and Sedj-1-M7 in ∆Sepi3k mutants alone. ∆Sepi3k mutants and WT (S. eturmiunum) strains were used as controls. a Eight transformants and control strains were cultured on PDA medium. The perithecia, asci and ascospores were photographed after growing for 6 weeks. b Eight transformants and control strains were cultured on CM medium. Conidia, conidiophores and conidiogenous cells were photographed at 4 weeks. c The nuclei distributions in the mycelia of all tested transformants and control strains were examined by DIC and fluorescence microscopy after staining by DAPI. Bar = 20 μm, 50 μm and 500 μm
Discussion
The role of ASF1 in sexual reproduction was first described for the ascomycete Sordaria macrospora (Gesing et al. 2012). Whether ASF1 acts in a conserved manner to control both asexual and sexual reproduction has not been addressed. In this report, we characterized the roles of ASF1 and its protein partners in controlling both asexual and sexual reproduction in S. eturmiunum. Until now, sexual reproduction had been described in a few filamentous fungal species (Coppin et al. 1997). Stemphylium as an emerging model in the Dothideomycetes (refs), and understanding whether the sexual process is similar among major ascomycete fungi. Stemphylium is an important genus in filamentous fungi, in which S. eturmiunum is a holotype species. S. eturmiunum has one copy ASF1 (SeASF1) which is more than 90% identical to the ASF1 gene in S. lycopersici, as well as nine other fungal species (Fig. S2). On the other hand, it shares less than 50% similarity with ASF1 in Schizosaccharomyces pombe. SeASF1 shows a conserved nuclear localization and prominent effects on sexual reproduction, suggesting conservation of function by comparison to S. macrospora (Gesing et al. 2012). Moreover, it was clear that SeASF1 manipulated both asexual and sexual reproduction in S. eturmiunum (Figs. 2, S6), but the levels of transcriptional and post-translational modification of five genes, such as Gα subunit, Mat1, Mat2, Ste2, and Ste3, related to sexual reproduction in MAPK signaling pathway were hardly changed in ∆Seasf1 mutants compared with WT strains (Figs. S7, S8). Thus, it was hypothesized that SeASF1 is not involved in the MAPK signaling pathway related to the fungi sexual mating (Chen et al. 2002; Saito 2010) and might utilize other signaling pathways to promote sexual mating in S. eturmiunum. To further investigate this hypothesis, we performed an RNA-seq analysis on ∆Seasf1 in contrast to WT-vegetative, and WT-sexual strains (Figs. S9, S10). The SeDJ-1 and other six genes (Table S4) were identified as possible candidates for cooperating with SeASF1 to modulate asexual and sexual development in S. eturmiunum. SeDJ-1 was found to interact with SeASF1 and to play a crucial role in asexual and sexual development in S. eturmiunum (Figs. S11, S13–S19).
Previous studies revealed that DJ-1 was involved in multiple biological functions in mammals (Hijioka et al. 2017; Scumaci et al. 2020; Mencke et al. 2021; Nakamura et al. 2021). However, DJ-1-based regulation of asexual and sexual differentiation in mammals, plants, and fungi was poorly understood. Here, we found that SeDJ-1 involved in asexual and sexual development in S. eturmiunum (Fig. 3) and further illuminated the mechanisms by which SeDJ-1 influences asexual and sexual development. These mechanisms were further supported by measuring transcriptional levels of Seasf1, SeH4, and Sedj-1 in corresponding to different knockout mutants of these three genes and by confirming the interactions between SeASF1 and SeH4, as well as between SeDJ-1 and SeASF1/SeH4 in vivo and in vitro, respectively (Figs. S20, 4). However, SeH4 did not influence asexual and sexual development in S. eturmiunum and did not involve in the regulation of asexual and sexual development by SeDJ-1 or SeASF1. Also, the SeDJ-1 can cooperate with SeASF1 to modulate both asexual and sexual phenotypes in S. eturmiunum, but the functions of SeDJ-1 compromise to SeASF1 (Fig. S21). These results reinforce previous results in the related U. botrytis which showed that mating type genes performed major roles in both asexual and sexual sporulation pathways (Wang et al. 2017).
Evidence showed that DJ-1 was an important component in the PI3K/AKT signaling pathway (Yang et al. 2005) and might bind with various other factors to activate a variety of biological processes (van der Brug et al. 2008; Vasseur et al. 2012). Multiple downstream proteins were reported in the PI3K/AKT signaling pathway in mammals (Sitaram et al. 2009; Wang et al. 2013; Vasseur et al. 2009; Xu et al. 2020), but only GSK3 (SeGSK3) was found in PI3K/AKT signaling pathway in S. eturmiunum. The expression levels of SeGSK3 were up-regulated in both ∆Sedj-1 and ∆Sepi3k mutants. However, SePI3K showed down-expression levels in the ∆Sedj-1 mutants, and the same expression trends of SeDJ-1 showed in the ∆Sepi3k mutants (Fig. S22). Thus, SeDJ-1 is likely an upstream component of the SePI3K and SeGSK3 modules in the PI3K/AKT signaling pathway of S. eturmiunum. SeDJ-1 also interacted with SePI3K and SeGSK3 in vivo and in vitro (Fig. S23). In all, SeGSK3 negatively regulated the expression of SeDJ-1 and SePI3K but did not contribute to the asexual and sexual reproduction, whereas SeDJ-1, a positive regulator of SePI3K, involved in PI3K/AKT signaling pathway to regulate asexual and sexual features in S. eturmiunum (Fig. 5).
To determine if and how SeDJ-1 was involved in asexual and sexual reproduction in S. eturmiunum, ∆Sedj-1 or ∆Sepi3k mutants were compared to their overexpression strains in terms of the activity levels of SeDJ-1 and SePI3K, as well as their effect on asexual and sexual activity. It was found that over-expression of SePI3K in ∆Sedj-1 strains could recover the asexual and sexual states. Thus, SeDJ-1 and SePI3K are not only two important components of the PI3K/AKT signaling pathway, but also perform the similar functions for regulating asexual and sexual development in S. eturmiunum. To further illuminate the mechanism of SeDJ-1 regulating asexual and sexual reproduction, seven truncations of SeDJ-1 were obtained. In our experiments, SeDJ-1-M6 was defined as a critical segment for interaction of SeDJ-1 with SePI3K and was shown to be an essential segment for asexual and sexual reproduction upon comparation of OESedj-1-M6 and OESedj-1-M7 (Fig. 5). Thus, SeDJ-1-M6 plays a critical role in the PI3K/AKT signaling pathway. A model of the process is shown in Fig. 6. It is predicted that SeASF1 coupled to SeH4 is translocated into the nucleus and then interact with SeDJ-1, in which SeASF1 stimulates asexual and sexual means alone. Thus, SeDJ-1 contributes to arouse SePI3K to modulate asexual and sexual states. Together, SeASF1 manipulates asexual and sexual reproduction in S. eturmiunum by SeDJ-1 perception of PI3K/AKT signaling pathway.
A model for SeASF1 regulates asexual and sexual reproduction in S. eturmiunum by SeDJ-1 stimulation of PI3K/AKT signaling pathway. SeASF1, a molecular chaperone, interacts with SeH4 and then translocates into nucleus through the nuclear pore. After getting into nucleus, the dimer of SeASF1-SeH4 modulates DNA replication and nucleosome assembly. SeASF1-SeH4 combination with SeDJ-1 modulates asexual and sexual reproduction. The mutual interactions emerge between SeDJ-1 and SePI3K and then each of them modulates asexual and sexual reproduction. SeASF1 does not interact with SePI3K which is not impact on SePI3K to control reproduction. In all, SeASF1 regulates asexual and sexual reproduction in S. eturmiunum upon SeDJ-1 to stimulate PI3K/AKT signaling pathway
In conclusion, our works have broadened the scope of ASF1 functions in filamentous ascomycetes and implicate DJ-1 as a major component for binding to PI3K/AKT pathway. These data further illuminate that ASF1 cross-talk DJ-1 and PI3K is required for both asexual and sexual reproductive pathways.
References
Avvakumov N, Nourani A, Côté J (2011) Histone chaperones: modulators of chromatin marks. Mol Cell 41:502–514
Bai J, Guo C, Sun W, Li M, Meng X, Yu Y, Jin Y, Tong D, Geng J, Huang Q, Qi J, Fu S (2012) DJ-1 may contribute to metastasis of non-small cell lung cancer. Mol Biol Rep 39:2697–2703
Bardwell L, Cook JG, Chang EC, Cairns BR, Thorner J (1996) Signaling in the yeast pheromone response pathway: specific and high-affinity interaction of the mitogen-activated protein (MAP) kinases Kss1 and Fus3 with the upstream MAP kinase kinase Ste7. Mol Cell Biol 16:3637–3650
Bayram Ö, Braus GH (2012) Coordination of secondary metabolism and development in fungi: the velvet family of regulatory proteins. FEMS Microbiol Rev 36:1–24
Bayram Ö, Bayram ÖS, Ahmed YL, Maruyama J, Valerius O, Rizzoli SO, Ficner R, Irniger S, Braus GH (2012) The Aspergillus nidulans MAPK module AnSte11-Ste50-Ste7-Fus3 controls development and secondary metabolism. PLoS Genet 8:e1002816
Bernardi-Wenzel J, Quecine MC, Azevedo JL, Pamphile JA (2016) Agrobacterium-mediated transformation of Fusarium proliferatum. Genet Mol Res 15:15027944
Blackwell M (2011) The fungi: 1, 2, 3 … 5.1 million species? Am J Bot 98:426–438
Bobrowicz P, Pawlak R, Correa A, Bell-Pedersen D, Ebbole DJ (2002) The Neurospora crassa pheromone precursor genes are regulated by the mating type locus and the circadian clock. Mol Microbiol 45:795–804
Böhm J, Hoff B, O’Gorman CM, Wolfers S, Klix V, Binger D, Zadra I, Kürnsteiner H, Pöggeler S, Dyer PS, Kück U (2013) Sexual reproduction and mating-type—mediated strain development in the penicillin-producing fungus Penicillium chrysogenum. Proc Natl Acad Sci USA 110:1476–1481
Bonifati V, Rizzu P, van Baren MJ, Schaap O, Breedveld GJ, Krieger E, Dekker MC, Squitieri F, Ibanez P, Joosse M, van Dongen JW, Vanacore N, van Swieten JC, Brice A, Meco G, van Duijn CM, Oostra BA, Heutink P (2003) Mutations in the DJ-1 gene associated with autosomal recessive early-onset parkinsonism. Science 299:256–259
Câmara MPS, O’Neill NR, van Berkum P (2002) Phylogeny of Stemphylium spp. based on ITS and glyceraldehyde-3-phosphate dehydrogenase gene sequences. Mycologia 94:660–672
Chen J, Chen J, Lane S, Liu H (2002) A conserved mitogen-activated protein kinase pathway is required for mating in Candida albicans. Mol Microbiol 46:1335–1344
Chen Y, Kang M, Lu W, Guo Q, Zhang B, Xie Q, Wu Y (2012) DJ-1, a novel biomarker and a selected target gene for overcoming chemoresistance in pancreatic cancer. J Cancer Res Clin Oncol 138:1463–1474
Coppin E, Debuchy R, Arnaise S, Picard M (1997) Mating types and sexual development in filamentous ascomycetes. Microbiol Mol Biol Rev 61:411–428
Crespo P, Xu N, Simonds WF, Gutkind JS (1994) Ras dependent activation of MAP kinase pathway mediated by G-protein beta gamma subunits. Nature 369:418–420
Dacks J, Roger AJ (1999) The first sexual lineage and the relevance of facultative sex. J Mol Evol 48:779–783
Dai YC, Cui BK, Si J, He SH, Hyde KD, Yuan HS, Liu XY, Zhou LW (2015) Dynamics of the worldwide number of fungi with emphasis on fungal diversity in China. Mycol Prog 14:62
Das C, Roy S, Namjoshi S, Malarkey CS, Jones DN, Kutateladze TG, Churchill ME, Tyler JK (2014) Binding of the histone chaperone ASF1 to the CBP bromodomain promotes histone acetylation. Proc Natl Acad Sci USA 111:E1072-1081
deVisser JAGM, Elena SF (2007) The evolution of sex: empirical insights into the roles of epistasis and drift. Nat Rev Genet 8:139–149
Dos Reis TF, Mellado L, Lohmar JM, Silva LP, Zhou JJ, Calvo AM, Goldman GH, Brown NA (2019) GPCR-mediated glucose sensing system regulates light-dependent fungal development and mycotoxin production. PLoS Genet 15:e1008419
Dyer PS, O’Gorman CM (2011) A fungal sexual revolution: Aspergillus and Penicillium show the way. Curr Opin Microbiol 14:649–654
Eitoku M, Sato L, Senda T, Horikoshi M (2008) Histone chaperones: 30 years from isolation to elucidation of the mechanisms of nucleosome assembly and disassembly. Cell Mol Life Sci 65:414–444
Engelman JA, Luo J, Cantley LC (2006) The evolution of phos-phatidylinositol 3-kinases as regulators of growth and metabolism. Nat Rev Genet 7:606–619
Errede B, Gartner A, Zhou Z, Nasmyth K, Ammerer G (1993) MAP kinase-related FUS3 from S. cerevisiae is activated by STE7 in vitro. Nature 362:261–264
Fu C, Coelho MA, David-Palma M, Priest SJ, Heitman J (2019) Genetic and genomic evolution of sexual reproduction: echoes from LECA to the fungal kingdom. Curr Opin Genet Dev 58–59:70–75
Fu G, Dai J, Li Z, Chen F, Liu L, Yi L, Teng Z, Quan C, Zhang L, Zhou T, Donkersley P, Song S, Shi Y (2020) The role of STAT3/p53 and PI3K-AKT-mTOR signaling pathway on DEHP-induced reproductive toxicity in pubertal male rat. Toxicol Appl Pharmacol 404:115151
Gesing S, Schindler D, Fränzel B, Wolters D, Nowrousian M (2012) The histone chaperone ASF1 is essential for sexual development in the filamentous fungus Sordaria macrospora. Mol Microbiol 84:748–765
Gong J, Zhang L, Zhang Q, Li X, Xia XJ, Liu YY, Yang QS (2018) Lentiviral vector-mediated SHC3 silencing exacerbates oxidative stress injury in nigral dopamine neurons by regulating the PI3K-AKT-FoxO signaling pathway in rats with parkinson’s disease. Cell Physiol Biochem 49:971–984
Groth A, Corpet A, Cook AJ, Roche D, Bartek J, Lukas J, Almouzni G (2007) Regulation of replication fork progression through histone supply and demand. Science 318:1928–1931
Hadany L, Comeron JM (2008) Why are sex and recombination so common? Ann N Y Acad Sci 1133:26–43
Han KH, Lee DB, Kim JH, Kim MS, Han KY, Kim WS, Park YS, Kim HB, Han DM (2003) Environmental factors affecting development of Aspergillus nidulans. J Microbiol 41:34–40
Hawksworth DL (2012) Global species numbers of fungi: are tropical studies and molecular approaches contributing to a more robust estimate? Biodivers Conserv 21:2425–2433
Hawksworth DL, Lücking R (2017) Fungal diversity revisited: 2.2 to 3.8 million species. Microbiol Spectr 5(4):101128
Herskowitz I (1995) MAPK kinase pathways in yeast: for mating and more. Cell 80:187–197
Hijioka M, Inden M, Yanagisawa D, Kitamura Y (2017) DJ-1/PARK7: a new therapeutic target for neurodegenerative disorders. Biol Pharm Bull 40:548–552
Hyde KD, Norphanphoun C, Chen J, Dissanayake AJ, Doilom M, Hongsanan S, Jayawardena RS, Jeewon R, Perera RH, Thongbai B, Wanasinghe DN, Wisitrassameewong K, Tibpromma S, Stadler M (2018) Thailand’s amazing diversity: up to 96% of fungi in northern Thailand are novel. Fungal Divers 93:215–239
Hyde KD, Xu JC, Rapior S, Jeewon R, Lumyong S, Niego AGT, Abeywickrama PD, Aluthmuhandiram JVS, Brahamanage RS, Brooks S, Chaiyasen A, Chethana KWT, Chomnunti P, Chepkirui C, Chuankid B, de Silva NI, Doilom M, Faulds C, Gentekaki E, Gopalan V, Kakumyan P, Harishchandra D, Hemachandran H, Hongsanan S, Karunarathna A, Karunarathna SC, Khan S, Kumla J, Jayawardena RS, Liu JK, Liu N, Luangharn T, Macabeo APG, Marasinghe DS, Meeks D, Mortimer PE, Mueller P, Nadir S, Nataraja KN, Nontachaiyapoom S, O’Brien M, Penkhrue W, Phukhamsakda C, Ramanan US, Rathnayaka AR, Sadaba RB, Sandargo B, Samarakoon BC, Tennakoon DS, Siva R, Sriprom W, Suryanarayanan TS, Sujarit K, Suwannarach N, Suwunwong T, Thongbai B, Thongklang N, Wei DP, Wijesinghe SN, Winiski J, Yan J, Yasanthika E, Stadler M (2019) The amazing potential of fungi: 50 ways we can exploit fungi industrially. Fungal Divers 97:1–136
Inderbitzin P, Harkness J, Turgeon BG, Berbee ML (2005) Lateral transfer of mating system in Stemphylium. Proc Natl Acad Sci USA 102:11390–11395
Izumitsu K, Yoshimi A, Kubo D, Morita A, Saitoh Y, Tanaka C (2009) The MAPKK kinase ChSte11 regulates sexual/asexual development, melanization, pathogenicity, and adaptation to oxidative stress in Cochliobolus heterostrophus. Curr Genet 55:439–448
Kawasaki L, Sánchez O, Shiozaki K, Aguirre J (2002) SakA MAP kinase is involved in stress signal transduction, sexual development and spore viability in Aspergillus nidulans. Mol Microbiol 45:1153–1163
Kück U, Pöggeler S (2009) Cryptic sex in fungi. Fungal Biol Rev 23:86–90
Le S, Davis C, Konopka JB, Sternglanz R (1997) Two new S-phase-specific genes from Saccharomyces cerevisiae. Yeast 13:1029–1042
Lee SC, Ni M, Li W, Shertz C, Heitman J (2010) The evolution of sex: a perspective from the fungal kingdom. Microbiol Mol Biol Rev 74:298–340
Lee YJ, Kim WI, Park TH, Bae JH, Nam HS, Cho SW, Choi YJ, Lee SH, Cho MK (2021) Upregulation of DJ-1 expression in melanoma regulates PTEN/AKT pathway for cell survival and migration. Arch Dermatol Res 313:583–591
Lennox ES (1955) Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1:190–206
Li L, Wright SJ, Krystofova S, Park G, Borkovich KA (2007) Heterotrimeric G protein signaling in filamentous fungi. Annu Rev Microbiol 61:423–452
Li Q, Zhou H, Wurtele H, Davies B, Horazdovsky B, Verreault A, Zhang Z (2008) Acetylation of histone H3 lysine 56 regulates replication-coupled nucleosome assembly. Cell 134:244–255
Lin X, Hull CM, Heitman J (2005) Sexual reproduction between partners of the same mating type in Cryptococcus neoformans. Nature 434:1017–1021
Lin CH, Choi A, Bennett RJ (2011) Defining pheromone-receptor signaling in Candida albicans and related asexual Candida species. Mol Biol Cell 22:4918–4930
Liu S, Dean RA (1997) G protein alpha subunit genes control growth, development, and pathogenicity of Magnaporthe grisea. Mol Plant Microbe Interact 10:1075–1086
Liu Y, Yang K, Qin Q, Lin G, Hu T, Xu Z, Wang S (2018) G Protein α subunit GpaB is required for asexual development, aflatoxin biosynthesis and pathogenicity by regulating cAMP signaling in Aspergillus flavus. Toxins 10:117
Liu R, Chen Y, Liu G, Li C, Song Y, Cao Z, Li W, Hu J, Lu C, Liu Y (2020) PI3K/AKT pathway as a key link modulates the multidrug resistance of cancers. Cell Death Dis 11:797
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−∆∆CT method. Methods 25:402–408
Lucas MT, Webster J (1964) Conidia of Pleospora scirpicola and P. valesiaca. Trans Br Mycol Soc 47:247–256
Lyu X, Shen C, Xie J, Fu Y, Jiang D, Hu Z, Tang L, Tang L, Ding F, Li K, Wu S, Hu Y, Luo L, Li Y, Wang Q, Li G, Cheng J (2015) A “footprint” of plant carbon fixation cycle functions during the development of a heterotrophic fungus. Sci Rep 5:12952
Maerz S, Ziv C, Vogt N, Helmstaedt K, Cohen N, Gorovits R, Yarden O, Seiler S (2008) The nuclear Dbf2-related kinase COT1 and the mitogen-activated protein kinases MAK1 and MAK2 genetically interact to regulate filamentous growth, hyphal fusion and sexual development in Neurospora crassa. Genetics 179:1313–1325
Mencke P, Boussaad I, Romano CD, Kitami T, Linster CL, Krüger R (2021) The role of DJ-1 in cellular metabolism and pathophysiological implications for parkinson’s disease. Cells 10:347
Messiaen S, Guiard J, Aigueperse C, Fliniaux I, Tourpin S, Barroca V, Allemand I, Fouchet P, Livera G, Vernet M (2016) Loss of the histone chaperone ASF1B reduces female reproductive capacity in mice. Reproduction 151:477–489
Metin B, Findley K, Heitman J (2010) The mating type locus (MAT) and sexual reproduction of Cryptococcus heveanensis: insights into the evolution of sex and sex-determining chromosomal regions in fungi. PLoS Genet 6:e10000961
Millar JB, Buck V, Wilkinson MG (1995) Pyp1 and Pyp2 PTPases dephosphorylate an osmosensing MAP kinase controlling cell size at division in fission yeast. Genes Dev 9:2117–2130
Min Y, Frost JM, Choi Y (2020) Gametophytic abortion in heterozygotes but not in homozygotes: implied chromosome rearrangement during T-DNA insertion at the ASF1 locus in Arabidopsis. Mol Cells 43:448–458
Mori T, Kawai-Toyooka H, Igawa T, Nozaki H (2015) Gamete dialogs in green lineages. Mol Plant 8:1442–1454
Mukherjee UA, Ong SB, Ong SG, Hausenloy DJ (2015) Parkinson’s disease proteins: novel mitochondrial targets for cardioprotection. Pharmacol Ther 156:34–43
Müller P, Aichinger C, Feldbrügge M, Kahmann R (1999) The MAP kinase kpp2 regulates mating and pathogenic development in Ustilago maydis. Mol Microbiol 34:1007–1017
Nair S (2020) Maternal control of gamete choice during fertilization. Int J Dev B Iol 64:175–180
Nakamura K, Sakai S, Tsuyama J, Nakamura A, Otani K, Kurabayashi K, Yogiashi Y, Masai H, Shichita T (2021) Extracellular DJ-1 induces sterile inflammation in the ischemic brain. PLoS Biol 19:e3000939
Naranjo-Ortiz MA, Gabaldón T (2019) Fungal evolution: major ecological adaptations and evolutionary transitions. Biol Rev 94:1443–1476
Neiman AM, Herskowitz I (1999) Reconstitution of a yeast protein kinase cascade in vitro: activation of the yeast MEK homologue STE7 by STE11. Proc Natl Acad Sci USA 91:3398–3402
Ni M, Feretzaki M, Sun S, Wang X, Heitman J (2011) Sex in fungi. Annu Rev Genet 45:405–430
Oh SE, Mouradian MM (2017) Regulation of signal transduction by DJ-1. Adv Exp Med Biol 1037:97–131
Patra K, Jana S, Sarkar A, Mandal DP, Bhattacharjee S (2019) The inhibition of hypoxia-induced angiogenesis and metastasis by cinnamaldehyde is mediated by decreasing HIF-1α protein synthesis via PI3K/AKT pathway. BioFactors 45:401–415
Peberdy JF (1980) Sexual reproduction in fungi. In: Developmental microbiology. Springer, Boston
Prado F, Cortés-Ledesma F, Aguilera A (2004) The absence of the yeast chromatin assembly factor Asf1 increases genomic instability and sister chromatid exchange. EMBO Rep 5:497–502
Ramesh MA, Malik SB, Logsdon JM Jr (2005) A phylogenomic inventory of meiotic genes: evidence for sex in Giardia and an early eukaryotic origin of meiosis. Curr Biol 15:185–191
Recht J, Tsubota T, Tanny JC, Diaz RL, Berger JM, Zhang X, Garcia BA, Shabanowitz J, Burlingame AL, Hunt DF, Kaufman PD, Allis CD (2006) Histone chaperone Asf1 is required for histone H3 lysine 56 acetylation, a modification associated with S phase in mitosis and meiosis. Proc Natl Acad Sci USA 103:6988–6993
Saito H (2010) Regulation of cross-talk in yeast MAPK signaling pathways. Curr Opin Microbiol 13:677–683
Sambrook JF, Russell DW (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor laboratory
Sanematsu F, Takami Y, Barman HK, Fukagawa T, Ono T, Shibahara KI, Nakayama T (2006) Asf1 is required for viability and chromatin assembly during DNA replication in vertebrate cells. J Biol Chem 281:13817–13827
Scumaci D, Olivo E, Fiumara CV, La Chimia M, De Angelis MT, Mauro S, Costa G, Ambrosio FA, Alcaro S, Agosti V, Costanzo FS, Cuda G (2020) DJ-1 proteoforms in breast cancer cells: the escape of metabolic epigenetic misregulation. Cells 9:1968
Shao P, Wang Y, Zhang M, Wen X, Zhang J, Xu Z, Hu M, Jiang J, Liu T (2019) The interference of DEHP in precocious puberty of females mediated by the hypothalamic IGF-1/PI3K/AKT/mTOR signaling pathway. Ecotoxicol Environ Saf 181:362–369
Simmons EG (1967) Typification of Alternaria, Stemphylium and Ulocladium. Mycologia 59:67–92
Simmons EG (1969) Perfect states of Stemphylium. Mycologia 60:1–26
Simmons EG (1989) Macrospora Fuckel (Pleosporales) and related anamorphs. Sydowia 41:314–329
Sitaram RT, Cairney CJ, Grabowski P, Keith WN, Hallberg B, Ljungberg B, Roos G (2009) The PTEN regulator DJ-1 is associated with hTERT expression in clear cell renal cell carcinoma. Int J Cancer 125:783–790
Srinivasan S, Ohsugi M, Liu Z, Fatrai S, Bernal-Mizrachi E, Permutt MA (2005) Endoplasmic reticulum stress-induced apoptosis is partly mediated by reduced insulin signaling through phosphatidylinositol 3-kinase/AKT and increased glycogen synthase kinase-3beta in mouse insulinoma cells. Diabetes 54:968–975
Stajich JE, Harris T, Brunk BP, Brestelli J, Fischer S, Harb OS, Kissinger JC, Li W, Nayak V, Pinney DF, Stoeckert CJ Jr, Roos DS (2012) FungiDB: an integrated functional genomics database for fungi. Nucleic Acids Res 40:675–681
Štorchová H, Hrdličková R, Chrtek J, Tetera M, Fitze D, Fehrer J (2000) An improved method of DNA isolation from plants collected in the field and conserved in saturated NaCl/CTAB solution. Taxon 49:79–84
Studt L, Humpf HU, Tudzynski B (2013) Signaling governed by G proteins and cAMP is crucial for growth, secondary metabolism and sexual development in Fusarium fujikuroi. PLoS ONE 8:e58185
Sutton A, Bucaria J, Osley MA, Sternglanz R (2001) Yeast asf1 protein is required for cell cycle regulation of histone gene transcription. Genetics 158:587–596
Taira T, Saito Y, Niki T, Iguchi-Ariga SM, Takahashi K, Ariga H (2004) DJ-1 has a role in antioxidative stress to prevent cell death. EMBO Rep 5:213–218
Taylor DL, Hollingsworth TN, McFarland JW, Lennon NJ, Nusbaum C, Ruess RW (2014) A first comprehensive census of fungi in soil reveals both hyperdiversity and fine-scale niche partitioning. Ecol Monogr 84:3–20
Teichert I, Steffens EK, Schnaß N, Fränzel B, Krisp C, Wolters DA, Kück U (2014) PRO40 is a scaffold protein of the cell wall integrity pathway, linking the MAP kinase module to the upstream activator protein kinase C. PLoS Genet 10:e1004582
van der Brug MP, Blackinton J, Chandran J, Hao LY, Lal A, Mazan-Mamczarz K, Martindale J, Xie C, Ahmad R, Thomas KJ, Beilina A, Gibbs JR, Ding J, Myers AJ, Zhan M, Cai H, Bonini NM, Gorospe M, Cookson MR (2008) RNA binding activity of the recessive parkinsonism protein DJ-1 supports involvement in multiple cellular pathways. Proc Natl Acad Sci USA 105:10244–10249
van Drogen F, Stucke VM, Jorritsma G, Peter M (2001) MAP kinase dynamics in response to pheromones in budding yeast. Nat Cell Biol 3:1051–1059
Vasseur S, Afzal S, Tardivel-Lacombe J, Park DS, Iovanna JL, Mak TW (2009) DJ-1/PARK7 is an important mediator of hypoxia-induced cellular responses. Proc Natl Acad Sci USA 106:1111–1116
Vasseur S, Afzal S, Tomasini R, Guillaumond F, Tardivel-Lacombe J, Mak TW, Iovanna JL (2012) Consequences of DJ-1 upregulation following p53 loss and cell transformation. Oncogene 31:664–670
Vitale S, Di Pietro A, Turrà D (2019) Autocrine pheromone signalling regulates community behaviour in the fungal pathogen Fusarium oxysporum. Nat Microbiol 4:1443–1449
Wallen RM, Perlin MH (2018) An overview of the function and maintenance of sexual reproduction in dikaryotic fungi. Front Microbiol 9:503
Wang Y, Liu W, He X, Zhou F (2013) Parkinson’s disease-associated DJ-1 mutations increase abnormal phosphorylation of tau protein through AKT/GSK-3β pathways. J Mol Neuro Sci 51:911–918
Wang Z, Li N, Li J, Dunlap JC, Trail F, Townsend JP (2016) The fast-evolving phy-2 gene modulates sexual development in response to light in the model fungus Neurospora crassa. Mbio 7:e02148
Wang Q, Wang S, Xiong CL, James TY, Zhang XG (2017) Mating-type genes of the anamorphic fungus Ulocladium botrytis affect both asexual sporulation and sexual reproduction. Sci Rep 7:7932
Wang S, Song C, Zhao L, Xu W, Li Z, Liu X, Zhang XG (2022) GTP binding protein Gtr1 cooperating with ASF1 regulates asexual development in Stemphylium eturmiunum. Int J Mol Sci 23:8355
Whiteway MS, Wu C, Leeuw T, Clark K, Fourest-Lieuvin A, Thomas DY, Leberer E (1995) Association of the yeast pheromone response G protein beta gamma subunits with the MAP kinase scaffold Ste5p. Science 269:1572–1575
Wilkins AS, Holliday R (2009) The evolution of meiosis from mitosis. Genetics 181:3–12
Willis KJ (2018) State of the World’s Fungi. Royal Botanic Gardens, Kew
Woudenberg JHC, Groenewald JZ, Binder M, Crous PW (2013) Alternaria redefined. Stud Mycol 75:171–212
Woudenberg JHC, Hanse B, van Leeuwen GCM, Groenewald JZ, Crous PW (2017) Stemphylium revisited. Stud Mycol 87:77–103
Xu F, Na L, Li Y, Chen L (2020) Roles of the PI3K/AKT/mTOR signalling pathways in neurodegenerative diseases and tumours. Cell Biosci 10:54
Yang Y, Gehrke S, Haque ME, Imai Y, Kosek J, Yang L, Beal MF, Nishimura I, Wakamatsu K, Ito S (2005) Inactivation of Drosophila DJ-1 leads to impairments of oxidative stress response and phosphatidylinositol 3-kinase/AKT signaling. Proc Natl Acad Sci USA 102:13670–13675
Yang C, Hou A, Yu C, Dai L, Wang W, Zhang K, Shao H, Ma J, Xu W (2018) Kanglaite reverses multidrug resistance of HCC by inducing apoptosis and cell cycle arrest via PI3K/AKT pathway. Onco Targets Ther 11:983–996
Yuan HS, Lu X, Dai YC, Hyde KD, Kan YH, Kušan I, He SH, Liu NG, Sarma VV, Zhao CL, Cui BK, Yousaf N, Sun GY, Liu SY, Wu F, Lin CG, Dayarathne MC, Gibertoni TB, Conceição LB, Garibay-Orijel R, Villegas-Ríos M, Salas-Lizana R, Wei TZ, Qiu JZ, Yu ZF, Phookamsak R, Zeng M, Paloi S, Bao DF, Abeywickrama PD, Wei DP, Yang J, Manawasinghe IS, Harishchandra D, Brahmanage RS, de Silva NI, Tennakoon DS, Karunarathna A, Gafforov Y, Pem D, Zhang SN, de Azevedo SALCM, Bezerra JDP, Dima P, Acharya K, Alvarez-Manjarrez J, Bahkali AH, Bhatt VK, Brandrud TE, Bulgakov TS, Camporesi E, Cao T, Chen YX, Chen YY, Devadatha B, Elgorban AM, Fan LF, Du X, Gao L, Gonçalves CM, Gusmão LFP, Huanraluek N, Jadan M, Jayawardena RS, Khalid AN, Langer E, Lima DX, de Lima-Júnior NC, de Lira CRS, Liu JK, Liu S, Lumyong S, Luo ZL, Matočec Z, Niranjan M, Oliveira-Filho JRC, Papp V, Pérez-Pazos E, Phillips AJL, Qiu PL, Ren YH, Castañeda Ruiz RF, Semwa KC, Soop K, de Souza CAF, Souza-Motta CM, Sun LH, Xie ML, Yao YJ, Zhao Q, Zhou LW (2020) Fungal diversity notes 1277–1386: taxonomic and phylogenetic contributions to fungal taxa. Fungal Divers 104:1–266
Zhang Y, Gong XG, Wang ZZ, Sun HM, Guo ZY, Gai C, Hu JH, Ma L, Li P, Chen NH (2016) Protective effects of DJ-1 medicated AKT phosphorylation on mitochondrial function are promoted by Da-Bu-Yin-Wan in 1-methyl-4-phenylpyridinium-treated human neuroblastoma SHSY5Y cells. J Ethnopharmacol 187:83–93
Zhang C, Ren X, Wang X, Wan Q, Ding K, Chen L (2020) FgRad50 regulates fungal development, pathogenicity, cell wall integrity and the DNA damage response in Fusarium graminearum. Front Microbiol 10:2970
Zhou X, Li G, Xu JR (2011) Efficient approaches for generating GFP fusion and epitope-tagging constructs in filamentous fungi. Methods Mol Biol 722:199–212
Zhou Y, Li S, Li J, Wang D, Li Q (2017) Effect of microRNA-135a on cell proliferation, migration, invasion, apoptosis and tumor angiogenesis through the IGF-1/PI3K/AKT signaling pathway in non-small cell lung cancer. Cell Physiol Biochem 42:1431–1446
Acknowledgements
We thank Minou Nowrousian (Ruhr-Universität Bochum) for providing the Sordaria macrospora strains. We thank Jingze Zhang (Zhejiang University) for the transcriptome analysis and Daohong Jiang (Huazhong Agricultural University) for providing the plasmids. We thank Beijing igeneCode Biotech Co., Ltd. for the support with the quantitative proteomics data analysis. We would like to thank Dr. Shannon Elliot at Michigan State University for his assistance with English language and grammatical editing. This work was supported by grants from the National Natural Science Foundation of China (31230001, U200220015).
Funding
Funding was provided by National Natural Science Foundation of China (Grant nos. 31230001, U200220015).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there is no conflict of interest.
Additional information
Handling Editor: Jian-Kui Liu.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, S., Liu, X., Xiong, C. et al. ASF1 regulates asexual and sexual reproduction in Stemphylium eturmiunum by DJ-1 stimulation of the PI3K/AKT signaling pathway. Fungal Diversity 123, 159–176 (2023). https://doi.org/10.1007/s13225-023-00528-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13225-023-00528-1