Abstract
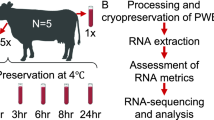
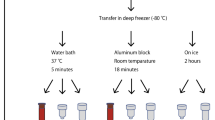
Whole blood is one of the most widely utilized human samples in biological research and is useful for analyzing the mechanisms of diverse bio-molecular phenomena. However, owing to its fluidic properties, whole blood is relatively unstable in the frozen state compared to other samples. Because RNA is structurally unstable, sample damage can severely affect RNA quality, thereby reducing its usability. This study aimed to assess the quality of RNA which was prepared from blood stored at different temperatures and times prior to freezing, as well as the effect of long-term freezing. The quality of the RNA derived from different blood samples was assessed by measuring the RNA integrity number (RIN) and RNA sequencing to identify differentially expressed genes (DEGs, |fold-change (FC)|> 1.5, p < 0.05, false discovery rate (FDR) < 0.05) between the differently prepared RNA samples. We found that improper sample handling critically influenced both RNA quality and gene expression patterns. By suggesting the consequences, this study emphasizes the importance of sample management to obtain reliable downstream application outcomes.



Similar content being viewed by others
References
Jeong, K.S., Kim, S., Kim, W.J., Kim, H.-C., Bae, J., Hong, Y.-C., Ha, M., Ahn, K., Lee, J.-Y., Kim, Y.: Cohort profile: beyond birth cohort study–The Korean CHildren’s ENvironmental health Study (Ko-CHENS). Environ. Res. 172, 358–366 (2019)
Elliott, P., Peakman, T.C.: The UK Biobank sample handling and storage protocol for the collection, processing and archiving of human blood and urine. Int. J. Epidemiol. 37(2), 234–244 (2008)
Shabihkhani, M., Lucey, G.M., Wei, B., Mareninov, S., Lou, J.J., Vinters, H.V., Singer, E.J., Cloughesy, T.F., Yong, W.H.: The procurement, storage, and quality assurance of frozen blood and tissue biospecimens in pathology, biorepository, and biobank settings. Clin. Biochem. 47(4–5), 258–266 (2014)
Opitz, L., Salinas-Riester, G., Grade, M., Jung, K., Jo, P., Emons, G., Ghadimi, B.M., Beißbarth, T., Gaedcke, J.: Impact of RNA degradation on gene expression profiling. BMC Biol. 3(1), 36 (2010)
Shen, Y., Li, R., Tian, F., Chen, Z., Lu, N., Bai, Y., Ge, Q., Lu, Z.: Impact of RNA integrity and blood sample storage conditions on the gene expression analysis. OncoTargets Ther. 11, 3573 (2018)
Augello, F.A., Rainen, L., Walenciak, M., Oelmüller, U., Wyrich, R., Bastian, H.: Method and device for collecting and stabilizing a biological sample. In. Google Patents (2003)
Beekman, J.M., Reischl, J., Henderson, D., Bauer, D., Ternes, R., Peña, C., Lathia, C., Heubach, J.F.: Recovery of microarray-quality RNA from frozen EDTA blood samples. J. Pharmacol. Toxicol. Methods 59(1), 44–49 (2009)
Kim, J.-H., Jin, H.-O., Park, J.-A., Chang, Y.H., Hong, Y.J., Lee, J.K.: Comparison of three different kits for extraction of high-quality RNA from frozen blood. PLoS ONE 3(1), 76 (2014)
Schroeder, A., Mueller, O., Stocker, S., Salowsky, R., Leiber, M., Gassmann, M., Lightfoot, S., Menzel, W., Granzow, M., Ragg, T.: The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 7(1), 1–14 (2006)
Kwon, N.Y., Ahn, J.J., Kim, J.-H., Kim, S.Y., Lee, J.H., Kwon, J.-H., Song, C.-S., Hwang, S.Y.: Rapid subtyping and pathotyping of avian influenza virus using Chip-based RT-PCR. BioChip J. 13(4), 333–340 (2019)
Livak, K.J., Schmittgen, T.D.: Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25(4), 402–408 (2001)
Yang, J., Sui, X., Wen, C., Pan, C., Zhu, Y., Zhang, J., Zhang, L.: A hemocompatible cryoprotectant inspired by freezing-tolerant plants. Colloids. Sufr. B. Biointerfaces 176, 106–114 (2019)
Koh, E.J., Hwang, S.Y.: Multi-omics approaches for understanding environmental exposure and human health. Mol. Cell. Toxicol. 15(1), 1–7 (2019)
Park, J., Kwon, S.O., Kim, S.-H., Kim, S.J., Koh, E.J., Won, S., Kim, W.J., Hwang, S.Y.: Methylation quantitative trait loci analysis in Korean exposome study. Mol. Cell. Toxicol. 16, 175–183 (2020)
Donohue, D.E., Gautam, A., Miller, S.-A., Srinivasan, S., Abu-Amara, D., Campbell, R., Marmar, C.R., Hammamieh, R., Jett, M.: Gene expression profiling of whole blood: a comparative assessment of RNA-stabilizing collection methods. PLoS ONE 14(10), e0223065 (2019)
Kågedal, B., Lindqvist, M., Farnebäck, M., Lenner, L., Peterson, C.: Failure of the PAXgeneTM Blood RNA System to maintain mRNA stability in whole blood. Clin. Chem. Lab. Med. 43(11), 1190–1192 (2005)
Acknowledgements
This study was supported by Korea Environmental Industry & Technology Institute (KEITI) through “the Environmental Health Action Program”, funded by Korea Ministry of Environment (MOE) (2017001360005)
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interests
All the authors declare that they have no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Koh, E.J., Yu, S.Y., Kim, S.H. et al. Understanding Confounding Effects of Blood Handling Strategies on RNA Quality and Transcriptomic Alteration Using RNA Sequencing. BioChip J 15, 187–194 (2021). https://doi.org/10.1007/s13206-021-00020-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13206-021-00020-5