Abstract
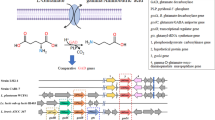
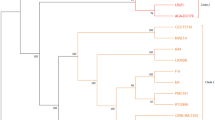
This study was conducted to investigate the γ-aminobutyric acid (GABA) production ability of 20 Lactobacillus and 25 Bifidobacterium strains which were previously isolated in our laboratory. Effect of initial pH, incubation time, monosodium glutamate (MSG), and pyridoxal-5′-phosphate (PLP) concentration for highest GABA production by two potent bacterial strains, Levilactobacillus brevis LAB6 and Limosilactobacillus fermentum LAB19 were optimized in the MRS media. A threefold increase in GABA production at an initial pH 4.0, incubation time of 120 h in medium supplemented with 3% MSG and 400 μM of PLP for LAB6 and 300 μM for LAB19 lead to the production of 19.67 ± 0.28 and 20.77 ± 0.14 g/L of GABA, respectively. Coculturing both strains under optimized conditions led to a GABA yield of 20.02 ± 0.17 g/L. Owing to potent anti-inflammatory activity in-vitro, as reported previously, and highest GABA production ability of LAB6 (MTCC 25662), its whole-genome sequencing and bioinformatics analysis was carried out for mining genes related to GABA metabolism. LAB6 harbored a complete glutamate decarboxylase (GAD) gene system comprising gadA, gadB, and gadC as well as genes responsible for the beneficial probiotic traits, such as for acid and bile tolerance and host adhesion. Comparative genomic analysis of LAB6 with 28 completely sequenced Levilactobacillus brevis strains revealed the presence of 95 strain-specific genes-families that was significantly higher than most other L. brevis strains.









Similar content being viewed by others
Data availability
The data sets generated during and/or analyzed during the current study are available from the corresponding author upon reasonable request. Whole genome sequencing data presented in this study are openly available in The National Centre for Biotechnology Information (NCBI).
References
Alcock BP, Raphenya AR, Lau TTY et al (2020) CARD 2020: antibiotic resistome surveillance with the comprehensive antibiotic resistance database. Nucleic Acids Res 48:D517–D525. https://doi.org/10.1093/nar/gkz935
Azcarate-Peril MA, Altermann E, Hoover-Fitzula RL et al (2004) Identification and inactivation of genetic loci involved with Lactobacillus acidophilus acid tolerance. Appl Environ Microbiol 70:5315–5322. https://doi.org/10.1128/AEM.70.9.5315-5322.2004
Aziz RK, Bartels D, Best AA et al (2008) The RAST Server: rapid annotations using subsystems technology. BMC Genom 9:75. https://doi.org/10.1186/1471-2164-9-75
Bauer KC, Huus KE, Finlay BB (2016) Microbes and the mind: emerging hallmarks of the gut microbiota-brain axis: emerging hallmarks of the gut microbiota-brain axis. Cell Microbiol 18:632–644. https://doi.org/10.1111/cmi.12585
Bhatia R, Singh S, Maurya R et al (2023) In vitro characterization of lactic acid bacterial strains isolated from fermented foods with anti-inflammatory and dipeptidyl peptidase-IV inhibition potential. Braz J Microbiol 54:293–309. https://doi.org/10.1007/s42770-022-00872-5
Chaudhari NM, Gupta VK, Dutta C (2016) BPGA- an ultra-fast pan-genome analysis pipeline. Sci Rep 6:24373. https://doi.org/10.1038/srep24373
Cho SY, Park MJ, Kim KM et al (2011) Production of high γ-aminobutyric acid (GABA) sour kimchi using lactic acid bacteria isolated from mukeunjee kimchi. Food Sci Biotechnol 20:403–408. https://doi.org/10.1007/s10068-011-0057-y
Chua J-Y, Koh MKP, Liu S-Q (2019) Gamma-aminobutyric acid. In: Sprouted Grains. Elsevier, pp 25–54
Couvin D, Bernheim A, Toffano-Nioche C et al (2018) CRISPRCasFinder, an update of CRISRFinder, includes a portable version, enhanced performance and integrates search for Cas proteins. Nucleic Acids Res 46:W246–W251. https://doi.org/10.1093/nar/gky425
Cui Y, Miao K, Niyaphorn S, Qu X (2020) Production of gamma-aminobutyric acid from lactic acid bacteria: a systematic review. Int J Mol Sci 21:995. https://doi.org/10.3390/ijms21030995
De Biase D, Tramonti A, Bossa F, Visca P (1999) The response to stationary-phase stress conditions in Escherichia coli: role and regulation of the glutamic acid decarboxylase system. Mol Microbiol 32:1198–1211. https://doi.org/10.1046/j.1365-2958.1999.01430.x
Demirbaş F, İspirli H, Kurnaz AA et al (2017) Antimicrobial and functional properties of lactic acid bacteria isolated from sourdoughs. Lebenson Wiss Technol 79:361–366. https://doi.org/10.1016/j.lwt.2017.01.067
Dhakal R, Bajpai VK, Baek K-H (2012) Production of gaba (γ - Aminobutyric acid) by microorganisms: a review. Braz J Microbiol 43:1230–1241. https://doi.org/10.1590/S1517-83822012000400001
Diana M, Quílez J, Rafecas M (2014) Gamma-aminobutyric acid as a bioactive compound in foods: a review. J Funct Foods 10:407–420. https://doi.org/10.1016/j.jff.2014.07.004
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. https://doi.org/10.1093/nar/gkh340
Edgar RC (2010) Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26:2460–2461. https://doi.org/10.1093/bioinformatics/btq461
Fan E, Huang J, Hu S et al (2012) Cloning, sequencing and expression of a glutamate decarboxylase gene from the GABA-producing strain Lactobacillus brevis CGMCC 1306. Ann Microbiol 62:689–698. https://doi.org/10.1007/s13213-011-0307-5
Gao D, Chang K, Ding G et al (2019) Genomic insights into a robust gamma-aminobutyric acid-producer Lactobacillus brevis CD0817. AMB Express 9:72. https://doi.org/10.1186/s13568-019-0799-0
Garcia-Gonzalez N, Bottacini F, van Sinderen D et al (2022) Comparative genomics of Lactiplantibacillus plantarum: Insights into probiotic markers in strains isolated from the human gastrointestinal tract and fermented foods. Front Microbiol 13:854266. https://doi.org/10.3389/fmicb.2022.854266
Gong L, Ren C, Xu Y (2019) Deciphering the crucial roles of transcriptional regulator GadR on gamma-aminobutyric acid production and acid resistance in Lactobacillus brevis. Microb Cell Fact 18:108. https://doi.org/10.1186/s12934-019-1157-2
Grant JR, Stothard P (2008) The CGView Server: a comparative genomics tool for circular genomes. Nucleic Acids Res 36:W181–W184. https://doi.org/10.1093/nar/gkn179
Higuchi T, Hayashi H, Abe K (1997) Exchange of glutamate and gamma-aminobutyrate in a Lactobacillus strain. J Bacteriol 179:3362–3364. https://doi.org/10.1128/jb.179.10.3362-3364.1997
Hoover DG (2014) Bifidobacterium. In: Encyclopedia of Food Microbiology. Elsevier, pp 216–222
Huang J, Mei L, Sheng Q et al (2007) Purification and characterization of glutamate decarboxylase of Lactobacillus brevis CGMCC 1306 isolated from fresh milk. Chin J Chem Eng 15:157–161. https://doi.org/10.1016/s1004-9541(07)60051-2
Huang J, Fang H, Gai Z-C et al (2018) Lactobacillus brevis CGMCC 1306 glutamate decarboxylase: crystal structure and functional analysis. Biochem Biophys Res Commun 503:1703–1709. https://doi.org/10.1016/j.bbrc.2018.07.102
Inoue K, Miyazaki Y, Unno K et al (2016) Stable isotope dilution HILIC-MS/MS method for accurate quantification of glutamic acid, glutamine, pyroglutamic acid, GABA and theanine in mouse brain tissues: HILIC-MS/MS assay of Glu analogs in mouse brain. Biomed Chromatogr 30:55–61. https://doi.org/10.1002/bmc.3502
Jitpakdee J, Kantachote D, Kanzaki H, Nitoda T (2022) Potential of lactic acid bacteria to produce functional fermented whey beverage with putative health promoting attributes. Lebenson Wiss Technol 160:113269. https://doi.org/10.1016/j.lwt.2022.113269
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30. https://doi.org/10.1093/nar/28.1.27
Krulwich TA, Sachs G, Padan E (2011) Molecular aspects of bacterial pH sensing and homeostasis. Nat Rev Microbiol 9:330–343. https://doi.org/10.1038/nrmicro2549
Lacroix N, St-Gelais D, Champagne CP, Vuillemard JC (2013) Gamma-aminobutyric acid-producing abilities of lactococcal strains isolated from old-style cheese starters. Dairy Sci Technol 93:315–327. https://doi.org/10.1007/s13594-013-0127-4
Li H, Cao Y (2010) Lactic acid bacterial cell factories for gamma-aminobutyric acid. Amino Acids 39:1107–1116. https://doi.org/10.1007/s00726-010-0582-7
Li H, Qiu T, Huang G, Cao Y (2010) Production of gamma-aminobutyric acid by Lactobacillus brevis NCL912 using fed-batch fermentation. Microb Cell Fact 9:85. https://doi.org/10.1186/1475-2859-9-85
Lin Q, Li D, Qin H (2017) Molecular cloning, expression, and immobilization of glutamate decarboxylase from Lactobacillus fermentum YS2. Electron J Biotechnol 27:8–13. https://doi.org/10.1016/j.ejbt.2017.03.002
Lu X, Xie C, Gu Z (2009) Optimisation of fermentative parameters for GABA enrichment by Lactococcus lactis. Czech J Food Sci 27:433–442. https://doi.org/10.17221/45/2009-cjfs
Lyu C, Zhao W, Peng C et al (2018) Exploring the contributions of two glutamate decarboxylase isozymes in Lactobacillus brevis to acid resistance and γ-aminobutyric acid production. Microb Cell Fact 17:180. https://doi.org/10.1186/s12934-018-1029-1
Mann EO, Paulsen O (2007) Role of GABAergic inhibition in hippocampal network oscillations. Trends Neurosci 30:343–349. https://doi.org/10.1016/j.tins.2007.05.003
Prévot T, Sibille E (2021) Altered GABA-mediated information processing and cognitive dysfunctions in depression and other brain disorders. Mol Psychiatry 26:151–167. https://doi.org/10.1038/s41380-020-0727-3
Rashmi D, Zanan R, John S, et al (2018) γ-Aminobutyric Acid (GABA): Biosynthesis, Role, Commercial Production, and Applications. In: Studies in Natural Products Chemistry. Elsevier, pp 413–452
Sanders JW, Leenhouts K, Burghoorn J et al (1998) A chloride-inducible acid resistance mechanism in Lactococcus lactis and its regulation. Mol Microbiol 27:299–310. https://doi.org/10.1046/j.1365-2958.1998.00676.x
Santos-Espinosa A, Beltrán-Barrientos LM, Reyes-Díaz R et al (2020) Gamma-aminobutyric acid (GABA) production in milk fermented by specific wild lactic acid bacteria strains isolated from artisanal Mexican cheeses. Ann Microbiol. https://doi.org/10.1186/s13213-020-01542-3
Sharma S, Singh S, Chaudhary V et al (2022) Isomaltooligosaccharides utilization and genomic characterization of human infant anti-inflammatory Bifidobacterium longum and Bifidobacterium breve strains. 3 Biotech 12:89. https://doi.org/10.1007/s13205-022-03141-2
Shi X, Chang C, Ma S et al (2017) Efficient bioconversion of L-glutamate to γ-aminobutyric acid by Lactobacillus brevis resting cells. J Ind Microbiol Biotechnol 44:697–704. https://doi.org/10.1007/s10295-016-1777-z
Singh S, Bhatia R, Khare P et al (2020) Anti-inflammatory Bifidobacterium strains prevent dextran sodium sulfate induced colitis and associated gut microbial dysbiosis in mice. Sci Rep 10:18597. https://doi.org/10.1038/s41598-020-75702-5
Siragusa S, De Angelis M, Di Cagno R et al (2007) Synthesis of gamma-aminobutyric acid by lactic acid bacteria isolated from a variety of Italian cheeses. Appl Environ Microbiol 73:7283–7290. https://doi.org/10.1128/AEM.01064-07
Sun Z, Harris HMB, McCann A et al (2015) Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera. Nat Commun 6:8322. https://doi.org/10.1038/ncomms9322
Tamura K, Stecher G, Kumar S (2021) MEGA11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 38:3022–3027. https://doi.org/10.1093/molbev/msab120
Tatusov RL, Galperin MY, Natale DA, Koonin EV (2000) The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res 28:33–36. https://doi.org/10.1093/nar/28.1.33
Teixeira JS, Seeras A, Sanchez-Maldonado AF et al (2014) Glutamine, glutamate, and arginine-based acid resistance in Lactobacillus reuteri. Food Microbiol 42:172–180. https://doi.org/10.1016/j.fm.2014.03.015
Thuy DTB, Nguyen A, Khoo KS et al (2020) Optimization of culture conditions for gamma-aminobutyric acid production by newly identified Pediococcus pentosaceus MN12 isolated from “mam nem”, a fermented fish sauce. Bioengineered 12:54–62. https://doi.org/10.1080/21655979.2020.1857626
Tramonti A, Visca P, De Canio M et al (2002) Functional characterization and regulation of gadX, a gene encoding an AraC/XylS-like transcriptional activator of the Escherichia coli glutamic acid decarboxylase system. J Bacteriol 184:2603–2613. https://doi.org/10.1128/JB.184.10.2603-2613.2002
Villegas JM, Brown L, Savoy de Giori G, Hebert EM (2016) Optimization of batch culture conditions for GABA production by Lactobacillus brevis CRL 1942, isolated from quinoa sourdough. Lebenson Wiss Technol 67:22–26. https://doi.org/10.1016/j.lwt.2015.11.027
Woraharn S, Lailerd N, Sivamaruthi BS et al (2015) Evaluation of factors that influence the L-glutamic and γ-aminobutyric acid production during Hericium erinaceus fermentation by lactic acid bacteria. CyTA - J Food 14:47–54. https://doi.org/10.1080/19476337.2015.1042525
Wu Q, Tun HM, Law Y-S et al (2017) Common distribution of gad operon in Lactobacillus brevis and its GadA contributes to efficient GABA Synthesis toward Cytosolic near-neutral pH. Front Microbiol 8:206. https://doi.org/10.3389/fmicb.2017.00206
Yao L-L, Cao J-R, Lyu C-J et al (2021) Food-grade γ-aminobutyric acid production by immobilized glutamate decarboxylase from Lactobacillus plantarum in rice vinegar and monosodium glutamate system. Biotechnol Lett 43:2027–2034. https://doi.org/10.1007/s10529-021-03164-4
Yogeswara IBA, Kittibunchakul S, Rahayu ES et al (2020) Microbial production and enzymatic biosynthesis of γ-aminobutyric acid (GABA) using Lactobacillus plantarum FNCC 260 isolated from Indonesian fermented foods. Processes (basel) 9:22. https://doi.org/10.3390/pr9010022
Zhu Z, Shi Z, Xie C et al (2019) A novel mechanism of Gamma-aminobutyric acid (GABA) protecting human umbilical vein endothelial cells (HUVECs) against H2O2-induced oxidative injury. Comp Biochem Physiol C Toxicol Pharmacol 217:68–75. https://doi.org/10.1016/j.cbpc.2018.11.018
Zhuang K, Jiang Y, Feng X et al (2018) Transcriptomic response to GABA-producing Lactobacillus plantarum CGMCC 1.2437T induced by L-MSG. PLoS One 13:e0199021. https://doi.org/10.1371/journal.pone.0199021
Acknowledgements
The authors would like to thank the Department of Biotechnology (DBT), Government of India and National Agri-Food Biotechnology Institute (NABI) for providing the research grant and the facilities. KK, KC, SRJ, and MB would like to acknowledge DBT project NERBPMC (BT/PR16088/NE/95/69/2015 NER-DBT) for providing the research grant. DeLCON (DBT e-library consortium) is highly acknowledged for providing access to the journals to NABI.
Funding
This research was funded by the Department of Biotechnology (DBT), Government of India, for the research grant given to the National Agri-Food Biotechnology Institute (NABI), Kanthi Kiran Kondepudi & Mahendra Bishnoi, Kanwaljit Chopra and Santa Ram Joshi.
Author information
Authors and Affiliations
Contributions
TM conducted the experiments and performed the whole-genome analysis and in-silico data analysis; RB isolation of Lactobacillus strains; KK, MB, KC, and SRJ provided the financial support for carrying out the research; KK, MB, and KC conceptualized, designed the experiments, edited the manuscript, and got funding to carry out the study. All authors have read and approved the final manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval
Not applicable.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Matta, T., Bhatia, R., Joshi, S.R. et al. GABA synthesizing lactic acid bacteria and genomic analysis of Levilactobacillus brevis LAB6. 3 Biotech 14, 62 (2024). https://doi.org/10.1007/s13205-024-03918-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-024-03918-7