Abstract
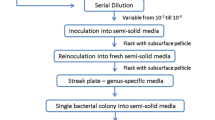
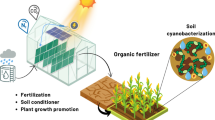
The metabolite profiles of two plant growth promoting cyanobacteria—Anabaena laxa and Calothrix elenkinii, which serve as promising biofertilizers, and biocontrol agents were generated to investigate their agriculturally beneficial activities. Preliminary biochemical analyses, in terms of total chlorophyll, total proteins, and IAA were highest at 14 days after inoculation (DAI). In A. laxa 20–25% higher values of reducing sugars, than C. elenkinii at both 14 and 21 DAI were recorded. Carbon and nitrogen assimilating enzyme activities—phosphoenol pyruvate carboxylase (PEPC), carbonic anhydrase (CA), and glutamine synthetase (GS) were highest at 14 DAI, albeit, nitrate reductase (NR) activity was higher by 0.73–0.84-fold at 21 DAI. Untargeted GC–MS (Gas chromatography–Mass spectrometric) analysis of metabolite profiles of 21d-old cyanobacterial cultures and characterization using NIST mass spectral library illustrated that A. laxa recorded highest number of metabolite hits in three chemical classes namely amino acid and peptides, nucleotides, nucleosides and analogues, besides other organic compounds. Based on the pathway analysis of identified metabolites, both A. laxa, and C. elenkinii were enriched in metabolites involved in aminoacyl-tRNA biosynthesis, and amino acid metabolism pathways, particularly lactose and glutamic acid, which are important players in plant–microbe interactions. Correlation-based metabolite network illustrated distinct and significant differences in the metabolic machinery of A. laxa and C. elenkinii, highlighting their novel identity and enrichment in C–N rich metabolites, as factors underlying their plant growth and soil fertility enhancing attributes, which make them valuable as inoculants.




Similar content being viewed by others
Availability of data and material
Raw metabolite data of cultures used in this study will be provided on reasonable request.
References
Abed RMM, Dobretsov S, Sudesh K (2009) Applications of cyanobacteria in biotechnology. J Appl Microbiol 106:1–12. https://doi.org/10.1111/j.1574-6976.2010.00234.x
Aebi H (1984) Catalase in vitro. Methods Enzymol 105:121–126. https://doi.org/10.1016/S0076-6879(84)05016-3
Azam R, Kothari R, Singh HM, Ahmad S, Sari A, Tyagi VV (2022) Cultivation of two Chlorella species in open sewage contaminated channel wastewater for biomass and biochemical profiles: comparative lab-scale approach. J Biotechnol 344:24–31. https://doi.org/10.1016/j.jbiotec.2021.11.006
Babu S, Bidyarani N, Chopra P, Monga D, Kumar R, Prasanna R, Kranthi S, Adak A, Saxena AK (2015) Evaluating microbe-plant interactions and varietal differences for enhancing biocontrol efficacy in root rot challenged cotton crop. Eur J Plant Pathol 142:345–362. https://doi.org/10.1007/s10658-015-0619-6
Badger MR, Price GD (1989) Carbonic anhydrase activity associated with the cyanobacterium Synechococcus PCC7942. Plant Physiol 89:51–60. https://doi.org/10.1104/pp.89.1.51
Bezrutczyk M, Yang J, Eom JS, Prior M, Sosso D, Hartwig T, Szurek B, Oliva R, Vera-Cruz C, White FF, Yang B (2018) Sugar flux and signalling in plant–microbe interactions. TPJ 93(4):675–685. https://doi.org/10.1111/tpj.13775
Bharti N, Pandey SS, Barnawal D, Patel VK, Kalra A (2016) Plant growth promoting rhizobacteria Dietzia natronolimnaea modulates the expression of stress responsive genes providing protection of wheat from salinity stress. Sci Rep 6:34768. https://doi.org/10.1038/srep34768
Bharti A, Prasanna R, Kumar G, Nain L, Rana A, Ramakrishnan B, Shivay YS (2021) Cyanobacterium-primed Chrysanthemum nursery improves the performance of the plant and soil quality. Biol Fertil Soils 57(1):87–105. https://doi.org/10.1007/s00374-020-01494-5
Bidyarani N, Prasanna R, Chawla G, Babu S, Singh R (2015) Deciphering the factors associated with the colonisation of rice plants by cyanobacteria. J Basic Microbiol 55:407–419. https://doi.org/10.1002/jobm.201400591
Chamizo S, Mugnai G, Rossi F, Certini G, De Philippis R (2018) Cyanobacteria inoculation improves soil stability and fertility on different textured soils: gaining insights for applicability in soil restoration. Front Environ Sci 6:49. https://doi.org/10.3389/fenvs.2018.00049
Choudhary M, Jetley UK, Khan MA, Zutshi S, Fatma T (2007) Effect of heavy metal stress on proline, malondialdehyde, and superoxide dismutase activity in the cyanobacterium Spirulina platensis-S5. Ecotoxicol Environ Saf 66:204–209. https://doi.org/10.1016/j.ecoenv.2006.02.002
Coppens J, Grunert O, Van Den Hende S, Vanhoutte I, Boon N, Haesaert G, De Gelder L (2016) The use of microalgae as a high-value organic slow-release fertilizer results in tomatoes with increased carotenoid and sugar levels. J Appl Phycol 28(4):2367–2377. https://doi.org/10.1007/s10811-015-0775-2
Dubois M, Gilles KA, Hamilton JK, Roberts PA, Smith F (1956) Colorimetric method for the determination of sugars and related substances. Anal Chem 28:350–356. https://doi.org/10.1021/ac60111a017
Ducat DC, Avelar-Rivas JA, Way JC, Silver PA (2012) Rerouting carbon flux to enhance photosynthetic productivity. Appl Environ Microbiol 78(8):2660–2668. https://doi.org/10.1128/aem.07901-11
Fa Y, Liang W, Cui H, Duan Y, Yang M, Gao J, Liu H (2015) Capillary ion chromatography–mass spectrometry for simultaneous determination of glucosylglycerol and sucrose in intracellular extracts of cyanobacteria. J Chromatogr B 1001:169–173. https://doi.org/10.1016/j.jchromb.2015.07.054
Forchhammer K, Selim KA (2020) Carbon/nitrogen homeostasis control in cyanobacteria. FEMS Microbiol Rev 44:33–53. https://doi.org/10.1093/femsre/fuz025
Garcia-Pichel F (2009) Cyanobacteria. In: Encyclopedia of microbiology. Elsevier Inc., pp 107–124. https://doi.org/10.1016/B978-012373944-5.00250-9
Gordon AS, Weber RP (1951) Colorimetric estimation of indole acetic acid. Plant Physiol 26:192–195
Gupta V, Natarajan C, Kumar K, Prasanna R (2011) Identification and characterization of endoglucanases for fungicidal activity in Anabaena laxa. J Appl Phycol 23:73–81. https://doi.org/10.1007/s10811-010-9539-1
Gupta V, Prasanna R, Chaudhary V, Nain L (2012) Biochemical, structural and functional characterization of two novel antifungal endoglucanases from Anabaena laxa. Biocatal Agric Biotechnol 1:338–347. https://doi.org/10.1016/j.bcab.2012.08.001
Hagemann M (2011) Molecular biology of cyanobacterial salt acclimation. FEMS Microbiol Rev 35(1):87–123. https://doi.org/10.1111/j.1574-6976.2010.00234.x
Herbert D, Phipps PJ, Strange RE (1971) Chemical analysis of microbial cells. In: Norris JR, Ribbons DW (eds) Methods in microbiology. Academic Press, New York, pp 209–344
Hu T, Oksanen K, Zhang W, Randell E, Furey A, Zhai G (2018) Analyzing feature importance for metabolomics using genetic programming. In: European Conference on Genetic Programming, Springer, Cham, pp 68–83. https://doi.org/10.1007/978-3-319-77553-1_5
Huang IS, Zimba PV (2019) Cyanobacterial bioactive metabolites—a review of their chemistry and biology. Harmful Algae 86:139–209. https://doi.org/10.1016/j.hal.2019.05.001
Hussain A, Hamayun M, Shah ST (2013) Root colonization and phytostimulation by phytohormones producing entophytic Nostoc sp. AH-12. Curr Microbiol 67:624–630. https://doi.org/10.1007/s00284-013-0408-4
Ito S, Osanai T (2020) Unconventional biochemical regulation of the oxidative pentose phosphate pathway in the model cyanobacterium Synechocystis sp. PCC 6803. Biochem J 477(7):1309–1321. https://doi.org/10.1042/BCJ20200038
Jablonsky J, Papacek S, Hagemann M (2016) Different strategies of metabolic regulation in cyanobacteria: from transcriptional to biochemical control. Sci Rep 6:33024. https://doi.org/10.1038/srep33024
Jin H, Ma H, Gan N, Wang H, Li Y, Wang L, Song I (2022) Non-targeted metabolomic profiling of filamentous cyanobacteria Aphanizomenon flos-aquae exposed to a concentrated culture filtrate of Microcystis aeruginosa. Harmful Algae 111:102–170. https://doi.org/10.1016/j.hal.2021.102170
Kokila V, Prasanna R, Saniya TK, Kumar A, Singh B (2023) Elevated CO2 modulates the metabolic machinery of cyanobacteria and valorizes its potential as a biofertilizer. Biocatal Agric Biotechnol 50:102716. https://doi.org/10.1016/j.bcab.2023.102716
Kundu A, Mishra S, Vadassery J (2018) Spodoptera litura-mediated chemical defense is differentially modulated in older and younger systemic leaves of Solanum lycopersicum. Planta 248:981–997. https://doi.org/10.1007/s00425-018-2953-3
Latifi A, Ruiz M, Zhang CC (2009) Oxidative stress in cyanobacteria. FEMS Microbiol Rev 33(2):258–278. https://doi.org/10.1111/j.1574-6976.2008.00134.x
Liu S, Hao H, Lu X, Zhao X, Wang Y, Zhang Y, Xie Z, Wang R (2017) Transcriptome profiling of genes involved in induced systemic salt tolerance conferred by Bacillus amyloliquefaciens FZB42 in Arabidopsis thaliana. Sci Rep 7(1):1–3. https://doi.org/10.1038/s41598-017-11308-8
Lorentz JF, Calijuri ML, Assemany PP, Alves WS, Pereira OG (2020) Microalgal biomass as a biofertilizer for pasture cultivation: plant productivity and chemical composition. J Clean Prod 276:124130. https://doi.org/10.1016/j.jclepro.2020.124130
Lowe RH, Evans HJ (1964) Preparation and some properties of a soluble nitrate reductase from Rhizobium japonicum. Biochim Biophys Acta 85:377–389. https://doi.org/10.1016/0926-6569(64)90301-3
Machado IM, Atsumi S (2012) Cyanobacterial biofuel production. J Biotechnol 162(1):50–56. https://doi.org/10.1016/j.jbiotec.2012.03.005
MacKinney G (1941) Absorption of light by chlorophyll solutions. J Biol Chem 140:315–322. https://doi.org/10.1016/S0021-9258(18)51320-X
Manjunath M, Prasanna R, Lata N, Dureja P, Singh R, Kumar A, Jaggi S, Kaushik BD (2010) Biocontrol potential of cyanobacterial metabolites against damping off disease caused by Pythium aphanidermatum in solanaceous vegetables. Arch Phytopathol Plant Prot 43(7):666–677. https://doi.org/10.1080/03235400802075815
Maqubela MP, Mnkeni PNS, Issa OM, Pardo MT, D’acqui LP (2009) Nostoc cyanobacterial inoculation in South African agricultural soils enhances soil structure, fertility, and maize growth. Plant Soil 315(1):79–92. https://doi.org/10.1007/s11104-008-9734-x
Miller GL (1959) Use of dinitrosalicylic acid reagent for determination of reducing sugar. Anal Chem 31:426–428. https://doi.org/10.1021/ac60147a030
Nakano Y, Asada K (1981) Hydrogen peroxide is scavenged by ascorbate-specific peroxidase in spinach chloroplasts. Plant Cell Physiol 22(5):867–880. https://doi.org/10.1093/oxfordjournals.pcp.a076232
Natarajan C, Prasanna R, Gupta V, Dureja P, Lata N (2012) Characterization of the fungicidal activity of Calothrix elenkinii using chemical analyses and microscopy. Appl Biochem Microbiol 48:53–57. https://doi.org/10.1134/S0003683812010115
Natarajan C, Gupta V, Kumar K, Prasanna R (2013) Molecular characterization of a fungicidal endoglucanase from the cyanobacterium Calothrix elenkinii. Biochem Genet 51:766–779. https://doi.org/10.1007/s10528-013-9605-x
Nishanth S, Prasanna R (2022) Untargeted GC-MS reveals differential regulation of metabolic pathways in cyanobacterium Anabaena and its biofilms with Trichoderma viride and Providencia sp. Curr Res Microb Sci 21:100174. https://doi.org/10.1016/j.crmicr.2022.100174
Oliver NJ, Rabinovitch-Deere CA, Carroll AL, Nozzi NE, Case AE, Atsumi S (2016) Cyanobacterial metabolic engineering for biofuel and chemical production. Curr Opin Chem Biol 35:43–50. https://doi.org/10.1016/j.cbpa.2016.08.023
Prasanna R, Lata N, Tripathi RM, Gupta V, Chaudhary V, Middha S, Joshi M, Ancha R, Kaushik BD (2008) Evaluation of fungicidal activity of extracellular filtrates of cyanobacteria - possible role of hydrolytic enzymes. J Basic Microbiol 48:186–194. https://doi.org/10.1002/jobm.200700199
Prasanna R, Gupta V, Natarajan C, Chaudhary V (2010) Bioprospecting for the genes involved in the production of chitosanases and microcystin-like compounds in Anabaena strains. World J Microbiol Biotechnol 26(4):717–724. https://doi.org/10.1007/s11274-009-0228-7
Prasanna R, Babu S, Rana A, Kabi SR, Chaudhary V, Gupta V, Kumar A, Shivay YS, Nain L, Pal RK (2013) Evaluating the establishment and agronomic proficiency of cyanobacterial consortia as organic options in wheat–rice cropping sequence. Exp Agric 49(3):416–434. https://doi.org/10.1017/S001447971200107X
Prasanna R, Ramakrishnan B, Ranjan K, Venkatachalam S, Kanchan A, Solanki P, Monga D, Shivay YS, Kranthi S (2016) Microbial inoculants with multifaceted traits suppress Rhizoctonia populations and promote plant growth in cotton. J Phytopathol 164(11–12):1030–1042. https://doi.org/10.1111/jph.12524
Prasannan CB, Jaiswal D, Davis R, Wangikar PP (2018) An improved method for extraction of polar and charged metabolites from cyanobacteria. PLoS ONE 13(10):e0204273. https://doi.org/10.1371/journal.pone.0204273
Renuka N, Guldhe A, Prasanna R, Singh P, Bux F (2018) Microalgae as multi-functional options in modern agriculture: current trends, prospects and challenges. Biotechnol Adv 36(4):1255–1273. https://doi.org/10.1016/j.biotechadv.2018.04.004
Roger PA, Tirol A, Santiago-Ardales S, Watanabe I (1986) Chemical composition of cultures and natural samples of N2-fixing blue-green algae from rice fields. Biol Fertil Soils 2:131–146. https://doi.org/10.1007/BF00257592
Rosier A, Medeiros FHV, Bais HP (2018) Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil 428:35–55. https://doi.org/10.1007/s11104-018-3679-5
Sarkar A, Rajarathinam R, Kumar PS, Rangasamy G (2022) Maximization of growth and lipid production of a toxic isolate of Anabaena circinalis by optimization of various parameters with mathematical modeling and computational validation. J Biotechnol 357:38–46. https://doi.org/10.1016/j.jbiotec.2022.08.001
Shapiro BM, Stadtman ER (1970) Glutamine synthetase (Escherichia coli). In: Tabor H, Tabor CW (eds) Methods in enzymology. Academic Press, New York, pp 910–922
Sharma V, Prasanna R, Hossain F, Muthusamy V, Nain L, Das S, Shivay YS, Kumar A (2020) Priming maize seeds with cyanobacteria enhances seed vigour and plant growth in elite maize inbreds. 3 Biotech 10:154. https://doi.org/10.1007/s13205-020-2141-6
Shinde S, Zhang X, Singapuri SP, Kalra I, Liu X, Morgan-Kiss RM, Wang X (2020) Glycogen metabolism supports photosynthesis start through the oxidative pentose phosphate pathway in cyanobacteria. Plant Physiol 182(1):507–517. https://doi.org/10.1104/pp.19.01184
Shivay YS, Prasanna R, Mandi S, Kanchan A, Simranjit K, Nayak S, Baral K, Sirohi MP, Nain L (2022) Cyanobacterial inoculation enhances nutrient use efficiency and grain quality of Basmati rice in the system of rice intensification (SRI). ACS Agric Sci Technol 2(4):742–753. https://doi.org/10.1021/acsagscitech.2c00030
Shukla J, Gulia U, Gupta H, Gupta K, Gogoi R, Kumar A, Mahawar H, Nishanth S, Saxena G, Singh AK, Nain L, Shivay YS, Prasanna R (2022) Harnessing cyanobacterium-fungal interactions to develop potting mixes for disease-free tomato nursery. Phytoparasitica. https://doi.org/10.1007/s12600-022-01011-4
Simranjit K, Kanchan A, Prasanna R, Ranjan K, Ramakrishnan B, Singh AK, Shivay YS (2019) Microbial inoculants as plant growth stimulating and soil nutrient availability enhancing options for cucumber under protected cultivation. World J Microbiol Biotechnol 35:51. https://doi.org/10.1007/s11274-019-2623-z
Stacey G, Tabita FR, Baalen CV (1977) Nitrogen and ammonia assimilation in the cyanobacteria: purification of glutamine synthetase from Anabaena sp. strain CA. J Bacteriol 132(2):596–603. https://doi.org/10.1128/jb.132.2.596-603.1977
Stanier RY, Cohen-Bazire G (1977) Phototrophic prokaryotes: the cyanobacteria. Annu Rev Microbiol 31:225–274. https://doi.org/10.1146/annurev.mi.31.100177.001301
Stirk WA, Ordog V, Staden JV, Jager K (2002) Cytokinins and auxin-like activity in Cyanophyta and microalgae. J Appl Phycol 14:215–221. https://doi.org/10.1023/A:1019928425569
Wu MX, Wedding RT (1985) Diurnal regulation of phosphoenolpyruvate carboxylase from Crassula. Plant Physiol 77(3):667–675. https://doi.org/10.1104/pp.77.3.667
Xia J, Wishart DS (2011) Web-based inference of biological patterns, functions and pathways from metabolomic data using MetaboAnalyst. Nat Protoc 6(6):743–760. https://doi.org/10.1038/nprot.2011.319
Acknowledgements
The authors are thankful to the Division of Microbiology, ICAR-Indian Agricultural Research Institute, New Delhi for the necessary facilities, and the Department of Science and Technology (DST), New Delhi for providing the DST-INSPIRE Fellowship (IF180455) to SN. The authors also acknowledge the Metabolomics Facility, NIPGR, and DBT grant (no. BT/INF/22/SP28268/2018) for funds towards the maintenance of GC/MS facility. SN is also grateful to Dr. Shobana Narayanasamy, Research Associate, Biocatalysts Laboratory, Tamil Nadu Agricultural University, Coimbatore, India for her guidance in correlation-based network analysis using Metscape application visualised using Cytoscape version 3.9.1. The authors are also thankful for receiving partial financial support from the Indian Council of Agricultural Research (ICAR) through the Network Project on Microorganisms “Application of Microorganisms in Agricultural and Allied Sectors” (AMAAS). The authors thank Dr. Lata Nain for her kind help in undertaking the language and scientific editing of the manuscript.
Funding
Department of Science and Technology (DST), New Delhi is thanked for providing the DST-INSPIRE Fellowship (IF180455) to SN, and the Indian Council of Agricultural Research (ICAR) for providing partial funds through the Network Project on Microorganisms “Application of Microorganisms in Agricultural and Allied Sectors” (AMAAS) to RP towards the analyses.
Author information
Authors and Affiliations
Contributions
NS: Conceptualization, Methodology, Experimentation, Investigation, Formal analysis, Writing-Original draft; VK: Experimentation, Investigation, Formal Analysis; RP: Methodology, Resources, Supervision, Formal analysis, Writing- Original draft, Review & Editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results. The authors report no commercial or proprietary interest in any product or concept discussed in this article.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Nishanth, S., Kokila, V. & Prasanna, R. Metabolite profiling of plant growth promoting cyanobacteria—Anabaena laxa and Calothrix elenkinii, using untargeted metabolomics. 3 Biotech 14, 35 (2024). https://doi.org/10.1007/s13205-023-03902-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-023-03902-7