Abstract
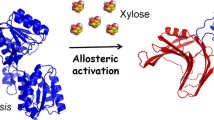
Development of chimeric enzymes by protein engineering can more efficiently contribute toward biomass conversion for bioenergy generation. Therefore, prior to experimental validation, a computational approach by modeling and molecular dynamic simulation can assess the structural and functional behavior of chimeric enzymes. In this study, a bifunctional chimera, CtXyn11A-BoGH43A comprising an efficient endoxylanase (CtXyn11A) from Clostridium thermocellum and xylosidase (BoGH43A) from Bacteroides ovatus was computationally designed and its binding and stability analysis with xylooligosaccharides were performed. The modeled chimera showed β-jellyroll fold for CtXyn11A and 5-bladed β-propeller fold for BoGH43A module. Stereo-chemical properties analyzed by Ramachandran plot showed 98.8% residues in allowed region, validating the modeled chimera. The catalytic residues identified by multiple sequence alignment were Glu94 and Glu184 for CtXyn11A and Asp229 and Glu384 for BoGH43A modules. CtXyn11A followed retaining-type, whereas BoGH43A enforced inverting-type of reaction mechanism during xylan hydrolysis as revealed by superposition and GH11 and GH43 familial analyses. Molecular docking studies showed binding energy, (ΔG) − 4.54 and − 4.18 kcal/mol for CtXyn11A and BoGH43A modules of chimera, respectively, with xylobiose, while − 3.94 and − 3.82 kcal/mol for CtXyn11A and BoGH43A modules of chimera, respectively, with xylotriose. MD simulation of CtXyn11A-BoGH43A complexed with xylobiose and xylotriose till 100 ns displayed stability by RMSD, compactness by Rg and conformational stability by SASA analyses. The lowered values of RMSF in active-site residues, Glu94, Glu184, Asp229, Asp335 and Glu384 confirmed the efficient binding of chimera with xylobiose and xylotriose. These results were in agreement with the earlier experimental studies on CtXyn11A releasing xylooligosaccharides from xylan and BoGH43A releasing d-xylose from xylooligosaccharides and xylobiose. The chimera showed stronger affinity in terms of total short-range interaction energy; − 190 and − 121 kJ/mol for with xylobiose and xylotriose, respectively. The bifunctional chimera, CtXyn11A-BoGH43A showed stability and integrity with xylobiose and xylotriose. The designed chimera can be constructed and applied for efficient biomass conversion.









Similar content being viewed by others
Data transparency
The authors declare that all data and materials as well as software application or custom code used in this study support their published claims and comply with the field standards.
Abbreviations
- GH:
-
Glycoside hydrolase
- MD:
-
Molecular dynamics
- RMSD:
-
Root-mean-square deviation
- Rg :
-
Radius of gyration
- SASA:
-
Solvent accessible surface area
- RMSF:
-
Root-mean-square-fluctuation
- MPD:
-
(4S)-2-methyl-2, 4-Pentanediol
- EDG:
-
1, 4-Di-oxy-1, 4-imino-l-Arabinitol
- XBB:
-
Xylobiose
- XTI:
-
Xylotriose
- XTT:
-
Xylotetraose
- XPT:
-
Xylopentaose
- Chimera:
-
CtXyn11A-BoGH43A
- CAZy:
-
Carbohydrate active enzyme database
References
Altschul SF, Warren G, Webb M, Eugene WM, David JL (1990) Basic local alignment search tool. J Mol Biol 215:403–410. https://doi.org/10.1016/S0022-2836(05)80360-2
Berendsen HJC, David S, Rudi D (1995) GROMACS: a message-passing parallel molecular dynamics implementation. Comput Phys Comm 91:43–56. https://doi.org/10.1016/0010-4655(95)00042-E
Binkowski TA, Andrzej J, Jie L (2005) Protein surface analysis for function annotation in high-throughput structural genomics pipeline. Protein Sci 14:2972–2981. https://doi.org/10.1110/ps.051759005
BIOVIA, Release 4 (2021) Discovery Studio, San Diego: Dassault Systèmes.
Colovos C, Todd OY (1993) Verification of protein structures: patterns of nonbonded atomic interactions. Protein Sci 2:1511–1519. https://doi.org/10.1002/pro.5560020916
Drula E, Garron ML, Dogan S, Lombard V, Henrissat B, Terrapon N (2022) The carbohydrate-active enzyme database: functions and literature. Nucleic Acids Res 50(D1):D571–D577. https://doi.org/10.1093/nar/gkab1045
Dundas J, Zheng O, Jeffery T, Andrew B, Yaron T, Jie L (2006) CASTp: computed atlas of surface topography of proteins with structural and topographical mapping of functionally annotated residues. Nucleic Acids Res 34:116–118. https://doi.org/10.1093/nar/gkl282
Eisenberg D, Roland L, James UB (1997) VERIFY3D: assessment of protein models with three-dimensional profiles. Methods Enzymol 277:396–404
Fan Z, Joshua RW, Ling Y (2009) Engineering of a multifunctional hemicellulase. Biotechnol Lett 31:751–757. https://doi.org/10.1007/s10529-009-9926-3
Gouet P, Xavier R, Emmanuel C (2003) ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Res 31:3320–3323. https://doi.org/10.1093/nar/gkg556
Gunsteren V, Wilfred F, Billeter SR, Eising AA, Hünenberger PH, Krüger PKHC, Mark AE, Scott WRP, Tironi IG (1996) Biomolecular simulation: the GROMOS96 manual and user guide. Vdf Hochschulverlag AG an Der ETH Zürich, Zürich 86:1–1044
Hess B, Henk B, Berendsen HJC, Johannes GEMF (1997) LINCS: a linear constraint solver for molecular simulations. J Comput Chem 18:1463–1472. https://doi.org/10.1002/(SICI)1096-987X(199709)18:12%3C1463::AID-JCC4%3E3.0.CO;2-H
Jamaldheen SB, Thakur A, Moholkar VS, Goyal A (2019) Enzymatic hydrolysis of hemicellulose from pretreated Finger millet (Eleusine coracana) straw by recombinant endo-1,4-β-xylanase and exo-1,4-β-xylosidase. Intl J Biol Macromol 15(135):1098–1106. https://doi.org/10.1016/j.ijbiomac.2019.06.010
Kelley LA, Stefans M, Christopher MY, Mark NW, Michael JES (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. http://www.nature.com/doifinder/10.1038/nprot.2015.053
Khaire KC, Sharma K, Thakur A, Moholkar VS, Goyal A (2021) Extraction and characterization of xylan from sugarcane tops as a potential commercial substrate. J Biosci Bioeng 131(6):647–654. https://doi.org/10.1016/j.jbiosc.2021.01.009
Koshland DE Jr (1953) Stereochemistry and the mechanism of enzymatic reactions. Biol Reviews 28(4):416–436. https://doi.org/10.1111/j.1469-185X.1953.tb01386.x
Lagaert S, Annick P, Christophe MC, Guido V (2014) β-Xylosidases and α-l-arabinofuranosidases: accessory enzymes for arabinoxylan degradation. Biotechnol Adv 32:316–332. https://doi.org/10.1016/j.biotechadv.2013.11.005
Laskowski RA, Malcolm WMA, David SM, Janet MT (1993) PROCHECK: a program to check the stereochemical quality of protein structures. J Appl Cryst 26:283–291. https://doi.org/10.1107/S0021889892009944
Lu P, Ming-Guang F (2008) Bifunctional enhancement of a β-glucanase-xylanase fusion enzyme by optimization of peptide linkers. Appl Microbiol Biotechnol 79:579–587. https://doi.org/10.1007/s00253-008-1468-4
Maki-Arvela P, Tapio S, Bjarne H, Stefan W, Dmitry YM (2011) Synthesis of sugars by hydrolysis of hemicelluloses-a review. Chem Rev 111:5638–5666. https://doi.org/10.1021/cr2000042
Malgas S, Mpho SM, Lithalethu M, Brett IP (2019) A mini review of xylanolytic enzymes with regards to their synergistic interactions during hetero-xylan degradation. World J Microbiol Biotechnol 35:1–13. https://doi.org/10.1007/s11274-019-2765-z
McKee LS, Maria JP, Artur R, Adam J, Richard JL, William SY, Kristian BRMK, Anders VN, Michael S, Harry JG, Jon MW (2012) Introducing endo-xylanase activity into an exo-acting arabinofuranosidase that targets side chains. Proc Natl Acad Sci 109:6537–6542. https://doi.org/10.1073/pnas.1117686109
Morris GM, Ruth H, William L, Michel FS, Richard KB, David SG, Arthur JO (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791. https://doi.org/10.1002/jcc.21256
Naidu DS, Shanganyane PH, Maya JJ (2018) Bio-based products from xylan: A review. Carbohydr Polym 179:28–41. https://doi.org/10.1016/j.carbpol.2017.09.064
Nath P, Dhillon A, Kumar K, Sharma K, Jamaldheen SB, Moholkar VS, Goyal A (2019) Development of bi-functional chimeric enzyme (CtGH1-L1-CtGH5-F194A) from endoglucanase (CtGH5) mutant F194A and β-1, 4-glucosidase (CtGH1) from Clostridium thermocellum with enhanced activity and structural integrity. Bioresour Technol 282:494–501. https://doi.org/10.1016/j.biortech.2019.03.051
Nath P, Sharma K, Kumar K, Goyal A (2020) Combined SAXS and computational approaches for structure determination and binding characteristics of Chimera (CtGH1-L1-CtGH5-F194A) generated by assembling β-glucosidase (CtGH1) and a mutant endoglucanase (CtGH5-F194A) from Clostridium thermocellum. Int J Biol Macromol 148:364–377. https://doi.org/10.1016/j.ijbiomac.2020.01.116
National Center for Biotechnology Information. PubChem Compound Summary for CID 25099191, Aprocitentan (2016) PubChem. https://pubchem.ncbi.nlm.nih.gov
Numan MT, Bhosle NB (2006) α-L-Arabinofuranosidases: the potential applications in biotechnology. J Ind Microbiol Biotechnol 33:247–260. https://doi.org/10.1007/s10295-005-0072-1
Parrinello M, Aneesur R (1980) Crystal structure and pair potentials: a molecular-dynamics study. Phys Rev Lett 45:1196. https://doi.org/10.1103/PhysRevLett.45.1196
Pettersen EF, Thomas DG, Conrad CH, Gregory SC, Daniel MG, Elaine CM, Thomas EF (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25:1605–1612. https://doi.org/10.1002/jcc.20084
Pourseif MM, Mitra Y, Mohammad A, Gholamali M, Ahmad N (2019) A multi-method and structure-based in silico vaccine designing against Echinococcus granulosus through investigating enolase protein. Bioimpacts 9:131. https://doi.org/10.15171/bi.2019.18
Ren J, Longping W, Xinjiao G, Changjiang J, Yu X, Xuebiao Y (2009) DOG 1.0: illustrator of protein domain structures. Cell Res 19:271–273. https://doi.org/10.1038/cr.2009.6
Rohman A, Niels VO, Ni NTP, Bauke WD (2018) Structural basis of product inhibition by arabinose and xylose of the thermostable GH43 β-1, 4-xylosidase from Geobacillus thermoleovorans IT-08. PLoS ONE 13:0196358. https://doi.org/10.1371/journal.pone.0196358
Saha BC (2003) Hemicellulose bioconversion. J Ind Microbiol Biotechnol 30:279–291. https://doi.org/10.1007/s10295-003-0049-x
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Schrodinger LLC (2019) The PyMOL molecular graphics system, Version 2.0
Schüttelkopf AW, Daan MFVA (2004) PRODRG: a tool for high-throughput crystallography of protein–ligand complexes. Acta Cryst 60:1355–1363. https://doi.org/10.1107/S0907444904011679
Sepulchro AGV, Vanessa OAP, Lorenzo B, Evandro AA, Simara SA, Igor P (2020) Transformation of xylan into value-added biocommodities using Thermobacillus composti GH10 xylanase. Carbohydr Polym 247:116714. https://doi.org/10.1016/j.carbpol.2020.116714
Sievers F, Desmond GH (2014) Clustal omega. Curr Protoc in Bioinformatics 48:3–13. https://doi.org/10.1002/0471250953.bi0313s48
Takita T, Kota N, Yuta K, Manami S, Kenji K, Naoki S, Bunzo M, Rie Y, Satoshi N, Kiyoshi Y (2019) Increase in the thermostability of GH11 xylanase XynJ from Bacillus sp. strain 41M–1 using site saturation mutagenesis. Enzyme Microb Technol 130:109363. https://doi.org/10.1016/j.enzmictec.2019.109363
Tamura K, Stecher G, Kumar S (2021) MEGA 11: molecular evolutionary genetics analysis version 11. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msab120
Wagschal K, Chamroeun H, Charles CL, Dominic WSW (2009) Biochemical characterization of a novel dual-function arabinofuranosidase/xylosidase isolated from a compost starter mixture. Appl Microbiol Biotechnol 81:855–863. https://doi.org/10.1007/s00253-008-1662-4
Wang J, Wei W, Peter AK, David A (2006) Case, automatic atom type and bond type perception in molecular mechanical calculations. J Mol Graphics Modell 25:247–260. https://doi.org/10.1016/j.jmgm.2005.12.005
Xin D, Xiang C, Peiyao W, Junhua Z (2019) Insight into the role of α-arabinofuranosidase in biomass hydrolysis: cellulose digestibility and inhibition by xylooligomers. Biotechnol Biofuels 12:1–11. https://doi.org/10.1186/s13068-019-1412-0
Yang J, Zhenggang H (2018) Understanding the positional binding and substrate interaction of a highly thermostable GH10 xylanase from Thermotoga maritima by molecular docking. Biomolecules 8:64. https://doi.org/10.3390/biom8030064
Zielkiewicz J (2005) Structural properties of water: comparison of the SPC, SPCE, TIP4P, and TIP5P models of water. J Chem Phys 123:104501. https://doi.org/10.1063/1.2018637
Acknowledgements
The authors thankfully acknowledge the financial support through a DBT-PAN-IIT Centre for Bioenergy: Phase II, Grant (No. BT/PR41982/PBD/26/822/2021), from Department of Biotechnology, Ministry of Science and Technology, New Delhi, India.
Funding
This study was funded by DBT-PAN-IIT Centre for Bioenergy: Phase II, Grant (No. BT/PR41982/PBD/26/822/2021).
Author information
Authors and Affiliations
Contributions
AG came up with the concept and devised the goals. SJ did the computational modeling, molecular docking and MD simulation of xylooligosaccharides with chimaera. PVG contributed in significance of the work, active site analysis, construction of the phylogeny and manuscript formatting. BC helped in simulation studies. The paper was written by SJ, PVG and AG.
Corresponding author
Ethics declarations
Conflicts of interest
The authors have declared that no potential conflicts of interest exist in relation to the content of this study.
Research involving human participants and/or animals
This research did not involve any human participants and/or animals on which experiments could be conducted.
Informed consent
This research did not involve any human participants whose consents could be taken for publication of this research-related data.
Additional information
Accession numbers: CtXyn11A: GenBank Accession—WP_014522591.1. BtGH43A: Uniprot ID: A7LXT8.1.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Ji, S., Gavande, P.V., Choudhury, B. et al. Computational design and structure dynamics analysis of bifunctional chimera of endoxylanase from Clostridium thermocellum and xylosidase from Bacteroides ovatus. 3 Biotech 13, 59 (2023). https://doi.org/10.1007/s13205-023-03482-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-023-03482-6