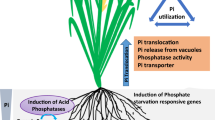
Abstract
Phosphorus (P) availability is determinant for crop productivity and, despite the sufficient amount of this nutrient in arable land, most of it remains insoluble, leading to the need of high fertilizer input. To cope with P scarcity forecasts and also for cropping costs alleviation, genotypes better adapted to promote P solubilization and absorption are required, especially for perennial crops. Coffee is one of these important perennial crops cultivated in soils with low P availability, and thus we aimed to study adaptive strategies to coffee genotypes in acquire phosphorus. In this study, we focused on rhizosphere phosphatase activity, a major characteristic related to P solubilization from organic pools. To explore the genetic basis of this characteristic, we firstly identified 29 Purple Acid Phosphatases (PAP) genes in C. canephora genome and selected five candidates with higher potential to encode secreted PAPs. We found that C. arabica genotypes have diverse profiles of rhizosphere phosphatase activity, as well as microbial biomass carbon, which we measured to explore the impact of the plant on microbiota and also to discriminate the phosphatase activity origin (plant or microorganism-derived). We selected two C. arabica cultivars with contrasting phosphatase activity and found that one PAP gene has a correlated gene expression profile with this characteristic. This work explores coffee adaptative responses to P starvation conditions, and the information provided can further contribute to breeding programs aiming better adapted genotypes for sustainable agriculture in face of current challenges.



Similar content being viewed by others
References
Abbasi-Vineh MA, Sabet MS, Karimzadeh G (2021) Identification and functional analysis of two purple acid phosphatases AtPAP17 and AtPAP26 involved in salt tolerance in Arabidopsis thaliana plant. Front Plant Sci 11:2326. https://doi.org/10.3389/fpls.2020.618716
Aleksieva P, Micheva-Viteva S (2000) Regulation of extracellular acid phosphatase biosynthesis by phosphates in proteinase producing fungus Humicola lutea 120–5. Enzyme Microbial Technol 27(8):570–575. https://doi.org/10.1016/S0141-0229(00)00237-4
Almeida EF et al (2007) Biomassa microbiana em sistema agroflorestal na zona da mata mineira. Cadernos Agroecol 2(2):739–742
Aon MA et al (2001) Spatio-temporal pattern of soil microbial and enzymatic activities in an agricultural soil. Appl Soil Ecol 18(3):239–254
Ascencio J (1994) Acid phosphatase as a diagnostic tool. Commun Soil Sci Plant Anal 25(9/10):1553–1564
Aziz T, Steffens D, Rahmatullah Schubert S (2011) Variation in phosphorus efficiency among Brassica cultivars II: changes in root morphology and carboxylate exudation. J Plant Nutr 34:2127–2138. https://doi.org/10.1080/01904167.2011.618573
Bernard M et al (2002) Characterization of a cell-wall acid phosphatase (PhoAp) in Aspergillus fumigatus. Microbiology 148(9):2819–2829. https://doi.org/10.1099/00221287-148-9-2819
Bieleski RL, Fergunson JB (1983) Physiology and metabolism of phosphate and its compounds. In: Pirson A, Zimmermann MH (eds) Encyclopedia of plant physiology: inorganic plant nutrition. Spring Verlag, Berlin, pp 422–4490
Carneiro MAC et al (2008) Atributos bioquímicos em dois solos de cerrado sob diferentes sistemas de manejo e uso. Pesquisa Agropecuária Trop 38(4):276–283
Chiou TJ, Lin SI (2011) Signaling network in sensing phosphate availability in plants. Annu Rev Plant Biol 62:185–206. https://doi.org/10.1146/annurev-arplant-042110-103849
Cordell D, Drangert JO, White S (2009) The story of phosphorus: global food security and food for thought. Glob Environ Change 19:292–305. https://doi.org/10.1016/j.gloenvcha.2008.10.009
Cruz CD (2013) Genes: a software package for analysis in experimental statistics and quantitative genetics. Acta Scient Agron 35(3):271–276. https://doi.org/10.4025/actasciagron.v35i3.21251
Deng S, Lu L, Li J et al (2020) Purple acid phosphatase 10c encodes a major acid phosphatase that regulates plant growth under phosphate-deficient conditions in rice. J Exp Bot 71(14):4321–4332. https://doi.org/10.1093/jxb/eraa179
Denoeud F, Carretero-Paulet L, Dereeper A et al (2014) The coffee genome provides insight into the convergent evolution of caffeine biosynthesis. Science 345(6201):1181–1184. https://doi.org/10.1126/science.1255274
Dick RP (1994) Soil enzymes activities as indicators of soil quality. In: Doran JW et al (eds) Defining soil quality for a sustainable environment. Soil Science Society of America, Madison, pp 107-124 (Special Publication number, 35)
Dick WA, Tabatabai MA (1993) Significance and potential uses of soil enzymes. In: Metting Junior FB (ed) Soil microbial ecology applications in agricultural and environmental management. M Dekker, New York, pp 95–127
Esposito E, Azevedo J (2004) Fungos: uma introdução à biologia, bioquímica e biotecnologia. Educs, Caxias do Sul
Fernandes LA et al (1998) Fósforo e atividade da fosfatase ácida em plantas de feijoeiro. Pesquisa Agropecuária Bras 33(5):769–778
Fernandes LA et al (2000) Frações de fósforo e atividade da fosfatase ácida em plantas de feijoeiro cultivadas em solos de várzea. Rev Bras Ciência Solo 24(3):561–571. https://doi.org/10.1590/S0100-06832000000300010
Fernandez DS, Ascencio J (1994) Acid phosphatase activity in bean and cowpea plants grown under phosphorus stress. J Plant Nutr 17(2/3):229–241. https://doi.org/10.1080/01904169409364723
Glaeser DF et al (2010) Biomassa microbiana do solo sob sistemas de manejo orgânico em cultivos de café. Ensaios Ciência 14(2):103–114
Gonçalves BX, Lima-Melo Y, dos Santos Maraschin F, Margis-Pinheiro M (2020) Phosphate starvation responses in crop roots: from well-known players to novel candidates. Environ Exp Bot 178:104162. https://doi.org/10.1016/j.envexpbot.2020.104162
Guimarães PTG et al (1999) Cafeeiro. In: Ribeiro AC, Guimarães PTG, Alvares VH (eds) Recomendações para uso de corretivos e fertilizantes em Minas Gerais: 5ª aproximação. Comissão de Fertilidade do Solo do Estado de Minas Gerais, Viçosa, MG, pp 289-302
Guimarães NF et al (2017) Biomassa e atividade microbiana do solo em diferentes sistemas de cultivo do cafeeiro. Rev Ciências Agrárias 40(1):34–44. https://doi.org/10.19084/RCA16041
Hidayat BJ, Eriksen NT, Wiebe MG (2006) Acid phosphatase production by Aspergillus niger N402A in continuous flow culture. FEMS Microbiol Lett 254(2):324–331. https://doi.org/10.1111/j.1574-6968.2005.00045.x
Hur YJ et al (2010) Molecular characterization of OsPAP2: transgenic expression of a purple acid phosphatase up-regulated in phosphate-deprived rice suspension cells. Biotechnol Lett 32(1):163–170. https://doi.org/10.1007/s10529-009-0131-1
Jakelaitis A et al (2008) Qualidade da camada superficial de solo sob mata, pastagens e áreas cultivadas. Pesquisa Agropecuária Trop 38(2):118–127
Kumar S, Stecher G, Tamura K (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol Biol Evol 33(7):1870–1874. https://doi.org/10.1093/molbev/msw054
Lammel DR et al (2015) Microbiological and faunal soil attributes of coffee cultivation under different management systems in Brazil. Braz J Biol 75(4):894–905. https://doi.org/10.1590/1519-6984.02414
Li D et al (2002) Purple acid phosphatases of Arabidopsis thaliana. Comparative analysis and differential regulation by phosphate deprivation. J Biol Chem 277(31):27772–27781. https://doi.org/10.1074/jbc.M204183200
Li C et al (2012) Identification of soybean purple acid phosphatase genes and their expression responses to phosphorus availability and symbiosis. Ann Bot. 109(1):275-285. https://doi.org/10.1093/aob/mcr246
Li R, An JP, You CX, Wang XF, Hao YJ (2020) Overexpression of MdPHR1 enhanced tolerance to phosphorus deficiency by increasing MdPAP10 transcription in apple (Malus× Domestica). J Plant Growth Regul 40:1753–1763. https://doi.org/10.1007/s00344-020-10225-x
Liang C et al (2010) Biochemical and molecular characterization of PvPAP3, a novel purple acid phosphatase isolated from common bean enhancing extracellular ATP utilization. Plant Physiol 152(2):854–865. https://doi.org/10.1104/pp.109.147918
Lu L, Qiu W, Gao W, Tyerman SD, Shou H, Wang C (2016) OsPAP10c, a novel secreted acid phosphatase in rice, plays an important role in the utilization of external organic phosphorus. Plant Cell Environ 39(10):2247–2259. https://doi.org/10.1111/pce.12794
Margalef O et al (2017) Global patterns of phosphatase activity in natural soils. Sci Rep 7(1337):1–13. https://doi.org/10.1038/s41598-017-01418-8
Marschner H, Rimmington G (1988) Mineral nutrition of higher plants. Plant Cell Environ 11:147–148
Mehra P, Pandey BK, Giri J (2017) Improvement in phosphate acquisition and utilization by a secretory purple acid phosphatase (OsPAP21b) in rice. Plant Biotechnol J 15(8):1054–1067. https://doi.org/10.1111/pbi.12699
Moreira FS, Siqueira JO (2006) Microbiologia bioquímica do solo, 2nd edn. Editora UFLA, Lavras
Nei M, Kumar S (2000) Molecular evolution and phylogenetics. Oxford University Press, Oxford
Novais RF, Neves JCL, Barros NF (1991) Ensaio em ambiente controlado. In: Oliveira AJ et al (coord). Métodos de pesquisa em fertilidade do solo. Embrapa-SEA, Brasília, pp 189–253
Partelli FL et al (2012) Chemical and microbiological soil characteristics under conventional and organic coffee production systems. Commun Soil Sci Plant Anal 43(5):847–864. https://doi.org/10.1080/00103624.2012.648470
Pfaffl MW et al (2004) Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: BestKeeper—Excel-based tool using pair-wise correlations. Biotechnol Lett 26(6):509–515. https://doi.org/10.1023/B:BILE.0000019559.84305.47
R Core Team (2020) R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. https://www.R-project.org/. Accessed 11 October 2021
Robinson WD et al (2012) The secreted purple acid phosphatase isozymes AtPAP12 and AtPAP26 play a pivotal role in extracellular phosphate-scavenging by Arabidopsis thaliana. J Exp Bot 63(18):6531–6542. https://doi.org/10.1093/jxb/ers309
Rössner H et al (1996) Indirect estimation of microbial biomass. In: Schinner F et al (eds) Methods in soil biology. Springer Verlag, Heidelberg, pp 47–75
Ruijter JM et al (2009) Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucl Acids Res 37(6):e45. https://doi.org/10.1093/nar/gkp045
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4(4):406–425. https://doi.org/10.1093/oxfordjournals.molbev.a040454
Silva FC, Basso LC (1993) Avaliação da atividade in vivo da fosfatase ácida da folha na diagnose da nutrição fosfórica em cana-de-açúcar. Rev Brasil Ciência Solo 17(3):371–375
Srivastava R, Parida AP, Chauhan PK, Kumar R (2020) Identification, structure analysis, and transcript profiling of purple acid phosphatases under Pi deficiency in tomato (Solanum lycopersicum L.) and its wild relatives. Int J Biol Macromol 165:2253–2266. https://doi.org/10.1016/j.ijbiomac.2020.10.080
Suen PK, Zhang S, Sun SSM (2015) Molecular characterization of a tomato purple acid phosphatase during seed germination and seedling growth under phosphate stress. Plant cell Rep 34(6):981–992. https://doi.org/10.1007/s00299-015-1759-z
Tabatabai MA, Bremner JM (1969) Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biol Biochem 1(4):301–307. https://doi.org/10.1016/0038-0717(69)90012-1
Thompson JD, Higgins DG, Gibson TJ (1994) Clustal W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res 22(22):4673–4680. https://doi.org/10.1093/nar/22.22.4673
Thumuluri V, Almagro Armenteros JJ, Johansen AR, Nielsen H, Winther O (2022) DeepLoc 2.0: multi-label subcellular localization prediction using protein language models. Nucleic Acids Res 50:W228–W234. https://doi.org/10.1093/nar/gkac278
Tian J, Liao H (2015) The role of intracellular and secreted purple acid phosphatases in plant phosphorus scavenging and recycling. In: Plaxton WC, Lambers H (eds) Annual plant reviews: phosphorus metabolism in plants. J. Wiley and Sons, Hoboken, pp 265–288. https://doi.org/10.1002/9781118958841.ch10
Tian J, Wang C, Zhang Q, He X, Whelan J, Shou H (2012) Overexpression of OsPAP10a, a root-associated acid phosphatase, increased extracellular organic phosphorus utilization in rice. J Integr Plant Biol 54(9):631–639. https://doi.org/10.1111/j.1744-7909.2012.01143.x
Tran HT, Hurley BA, Plaxton WC (2010) Feeding hungry plants: the role of purple acid phosphatases in phosphate nutrition. Plant Sci 179(1–2):14–27. https://doi.org/10.1016/j.plantsci.2010.04.005
Tran HT et al (2010) Biochemical and molecular characterization of AtPAP12 and AtPAP26: the predominant purple acid phosphatase isozymes secreted by phosphate-starved Arabidopsis thaliana. Plant Cell Environ 33(11):1789–1803. https://doi.org/10.1111/j.1365-3040.2010.02184.x
Tuomi JM et al (2010) Bias in the Cq value observed with hydrolysis probe based quantitative PCR can be corrected with the estimated PCR efficiency value. Methods 50(4):313–322. https://doi.org/10.1016/j.ymeth.2010.02.003
Vance CP (2001) Symbiotic nitrogen fixation and phosphorus acquisition. Plant nutrition in a world of declining renewable resources. Plant Physiol 127(2):390–397. https://doi.org/10.1104/pp.010331
Vance ED, Brookes PC, Jenkinson DS (1987) An extraction method for measuring soil microbial biomass. Soil Biol Biochem 19(6):703–707. https://doi.org/10.1016/0038-0717(87)90052-6
Venkidasamy B, Selvaraj D, Ramalingam S (2019) Genome-wide analysis of purple acid phosphatase (PAP) family proteins in Jatropha curcas L. Int J Biol Macromol 123:648–656. https://doi.org/10.1016/j.ijbiomac.2018.11.027
Vilela DJM, Coelho LS, Silva DRG et al (2021) Nutritional efficiency in phosphorus of arabica coffee genotypes. Coffee Sci 16:e161831. https://doi.org/10.25186/.v16i.1831
Wang L, Liu D (2018) Functions and regulation of phosphate starvation-induced secreted acid phosphatases in higher plants. Plant Sci 271:108–116. https://doi.org/10.1016/j.plantsci.2018.03.013
Wang L, Lu S, Zhang Y, Li Z, Du X, Liu D (2014) Comparative genetic analysis of Arabidopsis purple acid phosphatases AtPAP10, AtPAP12, and AtPAP26 provides new insights into their roles in plant adaptation to phosphate deprivation. J Integr Plant Biol 56(3):299–314. https://doi.org/10.1111/jipb.12184
Wu P et al (2013) Improvement of phosphorus efficiency in rice on the basis of understanding phosphate signaling and homeostasis. Curr Opin Plant Biol 16(2):205–212. https://doi.org/10.1016/j.pbi.2013.03.002
Xiao K et al (2006) Improved phosphorus acquisition and biomass production in Arabidopsis by transgenic expression of a purple acid phosphatase gene from M. truncatula. Plant Sci 170(2):191–202. https://doi.org/10.1016/j.plantsci.2005.08.001
Xie L, Shang Q (2018) Genome-wide analysis of purple acid phosphatase structure and expression in ten vegetable species. BMC Genomics 19(1):646. https://doi.org/10.1186/s12864-018-5022-1
Zhang Q et al (2011) Identification of rice purple acid phosphatases related to phosphate starvation signaling. Plant Biol 13(1):7–15. https://doi.org/10.1111/j.1438-8677.2010.00346.x
Zhu H, Qian W, Lu X, Li D, Liu X, Liu K, Wang D (2005) Expression patterns of purple acid phosphatase genes in Arabidopsis organs and functional analysis of AtPAP23 predominantly transcribed in flower. Plant Mol Biol 59(4):581–594. https://doi.org/10.1007/s11103-005-0183-0
Zhu S, Chen M, Liang C, Xue Y, Lin S, Tian J (2020) Characterization of purple acid phosphatase family and functional analysis of GmPAP7a/7b involved in extracellular ATP utilization in soybean. Front Plant Sci 11:661. https://doi.org/10.3389/fpls.2020.00661
Acknowledgments
To FAPEMIG, CNPq and Consórcio Pesquisa Café (Embrapa/Café) for financial support. To CAPES (Coordenação de Aperfeiçoamento de Pessoal de Nível Superior) for granting the scholarship. “This work was carried out with the support of the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior—Brasil (CAPES)—Financing Code 001”.
Author information
Authors and Affiliations
Contributions
DV, RP and TC carried out the experiments, analyzed the data and wrote the manuscript. LP, MAC and GC designed the research and edited the manuscript. JS designed the research, carried out the experiments and analyzed the data.
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Supplementary Information
Below is the link to the electronic supplementary material.





Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Vilela, D.J.M., Pinto, R.T., Cardoso, T.B. et al. Purple acid phosphatases in coffee—genome-wide identification and the transcriptional profile of selected members in roots upon Pi starvation. 3 Biotech 12, 335 (2022). https://doi.org/10.1007/s13205-022-03399-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03399-6