Abstract
A wealth of microarray and RNA-seq data for studying abiotic stress tolerance in rice exists but only limited studies have been carried out on multiple stress-tolerance responses and mechanisms. In this study, we identified 6657 abiotic stress-responsive genes pertaining to drought, salinity and heat stresses from the seedling stage microarray data of 83 samples and used them to perform unweighted network analysis and to identify key hub genes or master regulators for multiple abiotic stress tolerance. Of the total 55 modules identified from the analysis, the top 10 modules with 8–61 nodes comprised 239 genes. From these 10 modules, 10 genes common to all the three stresses were selected. Further, based on the centrality properties and highly dense interactions, we identified 7 intra-modular hub genes leading to a total of 17 potential candidate genes. Out of these 17 genes, 15 were validated by expression analysis using a panel of 4 test genotypes and a pair of standard check genotypes for each abiotic stress response. Interestingly, all the 15 genes showed upregulation under all stresses and in all the genotypes, suggesting that they could be representing some of the core abiotic stress-responsive genes. More pertinently, eight of the genes were found to be co-localized with the stress-tolerance QTL regions. Thus, in conclusion, our study not only provided an effective approach for studying abiotic stress tolerance in rice, but also identified major candidate genes which could be further validated by functional genomics for abiotic stress tolerance.





Similar content being viewed by others
Availability of data and material
All the input data and genes are available in the public domain except for the heat stress-responsive genes (HRG) and the appropriate references are given. HRG work is under publication and the gene list is available on request. Other information is available in the main manuscript or supplementary information.
Code availability
Not applicable.
References
Ahn H, Jung I, Shin SJ, Park J, Rhee S, Kim JK, Jung W, Kwon HB, Kim S (2017) Transcriptional network analysis reveals drought resistance mechanisms of AP2/ERF transgenic rice. Front Plant Sci 8:1044. https://doi.org/10.3389/fpls.2017.01044
Ambavaram MM, Basu S, Krishnan A, Ramegowda V, Batlang U, Rahman L, Baisakh N, Pereira A (2014) Coordinated regulation of photosynthesis in rice increases yield and tolerance to environmental stress. Nat Commun 5(1):1–4. https://doi.org/10.1038/ncomms6302
Amrine KC, Blanco-Ulate B, Cantu D (2015) Discovery of core biotic stress responsive genes in Arabidopsis by weighted gene co-expression network analysis. PloS One. https://doi.org/10.1371/journal.pone.0118731
Aya K, Suzuki G, Suwabe K, Hobo T, Takahashi H, Shiono K, Yano K, Tsutsumi N, Nakazono M, Nagamura Y, Matsuoka M (2011) Comprehensive network analysis of anther-expressed genes in rice by the combination of 33 laser microdissection and 143 spatiotemporal microarrays. PLoS One. https://doi.org/10.1371/journal.pone.0026162
Bader GD, Hogue CW (2003) An automated method for finding molecular complexes in large protein interaction networks. BMC Bioinform 4(1):2. https://doi.org/10.1186/1471-2105-4-2
Bindea G, Mlecnik B, Hackl H, Charoentong P, Tosolini M, Kirilovsky A, Fridman WH, Pagès F, Trajanoski Z, Galon J (2009) ClueGO: a cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25(8):1091–1093. https://doi.org/10.1093/bioinformatics/btp101
Carter H, Hofree M, Ideker T (2013) Genotype to phenotype via network analysis. Curr Opin Genet Dev 23(6):611–621. https://doi.org/10.1016/j.gde.2013.10.003
Childs KL, Davidson RM, Buell CR (2011) Gene coexpression network analysis as a source of functional annotation for rice genes. PloS One. https://doi.org/10.1371/journal.pone.0022196
Chinnusamy V, Schumaker K, Zhu JK (2004) Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J Exp Bot 55(395):225–236. https://doi.org/10.1093/jxb/erh005
Cohen SP, Leach JE (2019) Abiotic and biotic stresses induce a core transcriptome response in rice. Sci Rep 9(1):1–1. https://doi.org/10.1038/s41598-019-42731-8
Du Q, Campbell M, Yu H, Liu K, Walia H, Zhang Q, Zhang C (2019) Network-based feature selection reveals substructures of gene modules responding to salt stress in rice. Plant Direct 3(8):e00154. https://doi.org/10.1002/pld3.154
Ereful NC, Liu LY, Greenland A, Powell W, Mackay I, Leung H (2020) RNA-seq reveals differentially expressed genes between two indica inbred rice genotypes associated with drought-yield QTLs. Agronomy 10(5):621. https://doi.org/10.3390/agronomy10050621
Fukushima A, Kusano M, Redestig H, Arita M, Saito K (2009) Integrated omics approaches in plant systems biology. Curr Opin Chem Biol 13(5–6):532–538. https://doi.org/10.1016/j.cbpa.2009.09.022
Fukushima A, Kanaya S, Nishida K (2014) Integrated network analysis and effective tools in plant systems biology. Front Plant Sci 5:598. https://doi.org/10.3389/fpls.2014.00598
Gaiteri C, Ding Y, French B, Tseng GC, Sibille E (2014) Beyond modules and hubs: the potential of gene coexpression networks for investigating molecular mechanisms of complex brain disorders. Genes Brain Behav 13(1):13–24. https://doi.org/10.1111/gbb.12106
Gregoria GB, Senadhira D, Mendoza RD (1997) Screening rice for salinity tolerance. vol 22, IRRI discussion paper series. International Rice Research Institute
Habibpourmehraban F, Wu Y, Wu JX, Hamzelou S, Masoomi-Aladizgeh F, Kamath KS, Amirkhani A, Atwell BJ, Haynes PA (2022) Multiple abiotic stresses applied simultaneously elicit distinct responses in two contrasting rice cultivars. Int J Mol Sci 23(3):1739. https://doi.org/10.3390/ijms23031739
Hemamalini GS, Shashidhar HE, Hittalmani S (2000) Molecular marker assisted tagging of morphological and physiological traits under two contrasting moisture regimes at peak vegetative stage in rice (Oryza sativa L.). Euphytica 112(1):69–78. https://doi.org/10.1023/A:1003854224905
Kunihiro Y, Qian Q, Sato H, Teng S, Zeng DL, Fujimoto K, Zhu LH (2002) QTL analysis of sheath blight resistance in rice (Oryza sativa L.). Acta Genet Sin 29(1):50–5
Kurata N, Oryzabase Yamazaki Y (2006) An integrated biological and genome information database for rice. Plant Physiol 140(1):12–7. https://doi.org/10.1104/pp.105.063008
Lafitte HR, Price AH, Courtois B (2004) Yield response to water deficit in an upland rice mapping population: associations among traits and genetic markers. Theor Appl Genet 109(6):1237–46. https://doi.org/10.1007/s00122-004-1731-8
Li Y, Pearl SA, Jackson SA (2015) Gene networks in plant biology: approaches in reconstruction and analysis. Trends Plant Sci 20(10):664–675. https://doi.org/10.1016/j.tplants.2015.06.013
Liang T, Qing C, Liu P, Zou C, Yuan G, Pan G, Shen Y, Ma L (2022) Joint GWAS and WGCNA uncover the genetic control of calcium accumulation under salt treatment in maize seedlings. Physiol Plant 174(1):e13606. https://doi.org/10.1111/ppl.13606
Lima JM, Nath M, Dokku P, Raman KV, Kulkarni KP, Vishwakarma C, Sahoo SP, Mohapatra UB, Mithra SV, Chinnusamy V, Robin S (2015) Physiological, anatomical and transcriptional alterations in a rice mutant leading to enhanced water stress tolerance. AoB Plants. https://doi.org/10.1093/aobpla/plv023
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25(4):402–408. https://doi.org/10.1006/meth.2001.1262
Pandey S, Fartyal D, Agarwal A, Shukla T, James D, Kaul T, Negi YK, Arora S, Reddy MK (2017) Abiotic stress tolerance in plants: myriad roles of ascorbate peroxidase. Front Plant Sci 20(8):581. https://doi.org/10.3389/fpls.2017.00581
Plaimas K, Eils R, König R (2010) Identifying essential genes in bacterial metabolic networks with machine learning methods. BMC Syst Biol 4(1):56. https://doi.org/10.1186/1752-0509-4-56
Qu Y, Mu P, Zhang H, Chen CY, Gao Y, Tian Y, Wen F, Li Z (2008) Mapping QTLs of root morphological traits at different growth stages in rice. Genetica 133(2):187–200. https://doi.org/10.1007/s10709-007-9199-5
Ramkumar MK, Senthil Kumar S, Gaikwad K, Pandey R, Chinnusamy V, Singh NK, Singh AK, Mohapatra T, Sevanthi AM (2019) A novel stay-green mutant of rice with delayed leaf senescence and better harvest index confers drought tolerance. Plants 8(10):375. https://doi.org/10.3390/plants8100375
Ramu VS, Paramanantham A, Ramegowda V, Mohan-Raju B, Udayakumar M, Senthil-Kumar M (2016) Transcriptome analysis of sunflower genotypes with contrasting oxidative stress tolerance reveals individual-and combined-biotic and abiotic stress tolerance mechanisms. PloS One. https://doi.org/10.1371/journal.pone.0157522
Ravikiran KT, Krishnan SG, Vinod KK, Dhawan G, Dwivedi P, Kumar P, Bansal VP, Nagarajan M, Bhowmick PK, Ellur RK, Bollinedi H (2020) A trait specific QTL survey identifies NL44, a NERICA cultivar as a novel source for reproductive stage heat stress tolerance in rice. Plant Physiol Rep 25:664–676. https://doi.org/10.1007/s40502-020-00547-z
Samota MK, Sasi M, Awana M, Yadav OP, Amitha Mithra SV, Tyagi A, Kumar S, Singh A (2017) Elicitor-induced biochemical and molecular manifestations to improve drought tolerance in rice (Oryza sativa L.) through seed-priming. Front Plant Sci 8:934. https://doi.org/10.3389/fpls.2017.00934
Sandhu M, Sureshkumar V, Prakash C, Dixit R, Solanke AU, Sharma TR, Mohapatra T, Amitha Mithra SV (2017) RiceMetaSys for salt and drought stress responsive genes in rice: a web interface for crop improvement. BMC Bioinform 18(1):432. https://doi.org/10.1186/s12859-017-1846-y
Sandhu N, Singh V, Sihag MK (2019) Genomic footprints uncovering abiotic stress tolerance in rice. Advances in rice research for abiotic stress tolerance. Woodhead Publishing, pp 737–753
Scardoni G, Tosadori G, Faizan M, Spoto F, Fabbri F, Laudanna C (2014) Biological network analysis with CentiScaPe: centralities and experimental dataset integration. F1000Research. https://doi.org/10.12688/f1000research.4477.2
Sevanthi AM, Sinha SK, Sureshkumar V, Rani M, Saini MR, Kumari S, Kaushik M, Prakash C, Venkatesh K, Singh GP, Mohapatra T (2021) Integration of dual stress transcriptomes and major QTLs from a pair of genotypes contrasting for drought and chronic nitrogen starvation identifies key stress responsive genes in rice. Rice 14:49. https://doi.org/10.1186/s12284-021-00487-8
Shaik R, Ramakrishna W (2013) Genes and co-expression modules common to drought and bacterial stress responses in Arabidopsis and rice. PloS One. https://doi.org/10.1371/journal.pone.0077261
Shaik R, Ramakrishna W (2014) Machine learning approaches distinguish multiple stress conditions using stress-responsive genes and identify candidate genes for broad resistance in rice. Plant Physiol 164(1):481–495. https://doi.org/10.1104/pp.113.225862
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T (2003) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13(11):2498–2504. https://doi.org/10.1101/gr.1239303
Sharma R, De Vleesschauwer D, Sharma MK, Ronald PC (2013) Recent advances in dissecting stress-regulatory crosstalk in rice. Mol Plant 6(2):250–260. https://doi.org/10.1093/mp/sss147
Shi F, Dong Y, Wang M, Qiu D (2020) Transcriptomics analyses reveal that OsMIOX improves rice drought tolerance by regulating the expression of plant hormone and sugar related genes. Plant Biotechnol Rep 22:1–1. https://doi.org/10.1007/s11816-020-00608-7
Smita S, Katiyar A, Lenka SK, Dalal M, Kumar A, Mahtha SK, Yadav G, Chinnusamy V, Pandey DM, Bansal KC (2020) Gene network modules associated with abiotic stress response in tolerant rice genotypes identified by transcriptome meta-analysis. Funct Integr Genomic 20(1):29–49. https://doi.org/10.1007/s10142-019-00697-w
Suratanee A, Chokrathok C, Chutimanukul P, Khrueasan N, Buaboocha T, Chadchawan S, Plaimas K (2018) Two-state co-expression network analysis to identify genes related to salt tolerance in Thai rice. Genes 9(12):594. https://doi.org/10.3390/genes9120594
Suzuki N, Rivero RM, Shulaev V, Blumwald E, Mittler R (2014) Abiotic and biotic stress combinations. New Phytol 203(1):32–43. https://doi.org/10.1111/nph.12797
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M (2015) STRING v10: protein–protein interaction networks, integrated over the tree of life. Nucleic Acids Res 43(D1):D447–D452. https://doi.org/10.1093/nar/gku1003
Takehisa H, Sato Y, Antonio B, Nagamura Y (2015) Coexpression network analysis of macronutrient deficiency response genes in rice. Rice 8(1):24. https://doi.org/10.1186/s12284-015-0059-0
Tan M, Cheng D, Yang Y, Zhang G, Qin M, Chen J, Chen Y, Jiang M (2017) Co-expression network analysis of the transcriptomes of rice roots exposed to various cadmium stresses reveals universal cadmium-responsive genes. BMC Plant Biol 17(1):194. https://doi.org/10.1186/s12870-017-1143-y
Thomson MJ, de Ocampo M, Egdane J, Rahman MA, Sajise AG, Adorada DL, Tumimbang-Raiz E, Blumwald E, Seraj ZI, Singh RK, Gregorio GB (2010) Characterizing the Saltol quantitative trait locus for salinity tolerance in rice. Rice 3(2):148–160. https://doi.org/10.1007/s12284-010-9053-8
Van Dam S, Vosa U, van der Graaf A, Franke L, de Magalhaes JP (2018) Gene co-expression analysis for functional classification and gene–disease predictions. Brief Bioinfo 19(4):575–592. https://doi.org/10.1093/bib/bbw139
Vemanna RS, Bakade R, Bharti P, Kumar MK, Sreeman SM, Senthil-Kumar M, Makarla U (2019) Cross-talk signaling in rice during combined drought and bacterial blight stress. Front Plant Sci 10:193. https://doi.org/10.3389/fpls.2019.00193
Venuprasad R, Bool ME, Quiatchon L, Atlin GN (2012) A QTL for rice grain yield in aerobic environments with large effects in three genetic backgrounds. Theor Appl Genet 124(2):323–332. https://doi.org/10.1007/s00122-011-1707-4
Vikram P, Swamy BP, Dixit S, Ahmed HU, Teresa Sta Cruz M, Singh AK, Kumar A (2011) qDTY 11, a major QTL for rice grain yield under reproductive-stage drought stress with a consistent effect in multiple elite genetic backgrounds. BMC Genet 12(1):1–5. https://doi.org/10.1186/1471-2156-12-89
Wilkins O, Hafemeister C, Plessis A, Holloway-Phillips MM, Pham GM, Nicotra AB, Gregorio GB, Jagadish SK, Septiningsih EM, Bonneau R, Purugganan M (2016) EGRINs (environmental gene regulatory influence networks) in rice that function in the response to water deficit, high temperature, and agricultural environments. Plant Cell 28(10):2365–2384. https://doi.org/10.1105/tpc.16.00158
Xia L, Zou D, Sang J, Xu X, Yin H, Li M, Wu S, Hu S, Hao L, Zhang Z (2017) Rice expression database (RED): an integrated RNA-Seq-derived gene expression database for rice. J Genet Genom 44(5):235–41. https://doi.org/10.1016/j.jgg.2017.05.003
Yonemaru JI, Yamamoto T, Fukuoka S, Uga Y, Hori K, Yano M (2010) Q-TARO: QTL annotation rice online database. Rice 3(2–3):194–203. https://doi.org/10.1007/s12284-010-9041-z
Yue B, Xiong L, Xue W, Xing Y, Luo L, Xu C (2005) Genetic analysis for drought resistance of rice at reproductive stage in field with different types of soil. Theor Appl Genet 111(6):1127–1136. https://doi.org/10.1007/s00122-005-0040-1
Zheng S, Liu S, Feng J, Wang W, Wang Y, Yu Q, Liao Y, Mo Y, Xu Z, Li L, Gao X (2021) Overexpression of a stress response membrane protein gene OsSMP1 enhances rice tolerance to salt, cold and heavy metal stress. Environ Exp Bot 1(182):104327. https://doi.org/10.1016/j.envexpbot.2020.104327
Zhu M, Xie H, Wei X, Dossa K, Yu Y, Hui S, Tang G, Zeng X, Yu Y, Hu P, Wang J (2019) WGCNA analysis of salt-responsive core transcriptome identifies novel hub genes in rice. Genes 10(9):719. https://doi.org/10.3390/genes10090719
Funding
The financial assistance to carry out this work was obtained from the ICAR-CABin, a network project run by Indian council of agricultural research-Centre for agricultural bioinformatics and Department of Biotechnology (DBT), Government of India, through BT/PR10787/AGIII/103/883/2014.
Author information
Authors and Affiliations
Contributions
Conceptualization, MKR and AMS; Data curation, SSK; Facilities: NKS; Execution of plant stress studies and expression analysis, MKR, EM and VJ; Bioinformatics analyses, VS; Bioinformatics guidance, AUS; Material Resources, SG, MR and AKS; Overall guidance and supervision, AMS; Manuscript preparation – MKR and AMS. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethical approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
All the authors give their consent for publication of the research article to the journal, 3 Biotech.
Supplementary Information
Below is the link to the electronic supplementary material.
13205_2022_3182_MOESM1_ESM.tif
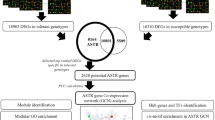
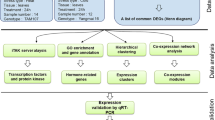
Supplementary file1 Workflow of bioinformatics analysis performed to identify candidate genes for multiple abiotic stress tolerance in rice (TIF 1359 KB)
Rights and permissions
About this article
Cite this article
Ramkumar, M.K., Mulani, E., Jadon, V. et al. Identification of major candidate genes for multiple abiotic stress tolerance at seedling stage by network analysis and their validation by expression profiling in rice (Oryza sativa L.). 3 Biotech 12, 127 (2022). https://doi.org/10.1007/s13205-022-03182-7
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-022-03182-7