Abstract
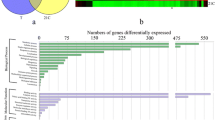
The goal of this study was to provide quantitative data on the catechin contents and underlying molecular regulatory mechanisms in cucumber during fruit development. The dynamic changes in the total catechin contents and RNA-seq-based transcriptome profiling of the flesh and peel of the cucumber cultivar ‘YanBai’, which is strongly astringent, were examined at three key developmental stages 3, 6 and 9 days post-pollination. The total catechin content decreased as cucumber fruit developed and was significantly lower in the flesh than in the peel. In total, 5092 and 4004 genes were found to be differently expressed in the peel and flesh, respectively. Based on a functional annotation, eight structural genes encode enzymes involved in the catechin biosynthesis pathway. Three genes encoding 4-coumarate-CoA ligases, two genes encoding chalcone isomerases, two genes encoding dihydroflavonol-4-reductase and one gene each encoding a phenylalanine ammonia-lyase, flavanone 3-hydroxylase and cinnamate 4-hydroxylase were identified as affecting the catechin content of cucumber. The transcriptome data also revealed the significance of transcription factors, including WD40-repeat proteins, MYB and bHLH, in regulating catechin biosynthesis. These findings help increase our understanding of the molecular mechanisms controlling catechin biosynthesis and astringency development in cucumber fruit.




Similar content being viewed by others
Data availability
The data generated or analyzed during this study are included in this published article, its supplementary information files, and publicly available repositories. The raw RNA-seq reads have been deposited in NCBI Gene Expression Omnibus under accession GSE112666.
References
Ando K, Carr K, Grumet R (2012) Transcriptome analyses of early cucumber fruit growth identifies distinct gene modules associated with phases of development. BMC Genom 13:518
Baldi P, Moser M, Brilli M, Vrhovsek U, Pindo M, Si-Ammour A (2017) Fine-tuning of the flavonoid and monolignol pathways during apple early fruit development. Planta 245:1021–1035
Baudry A, Heim M, Dubreucq B, Caboche M, Weisshaar B, Lepiniec L (2004) TT2, TT8, and TTG1 synergistically specify the expression of BANYULS and proanthocyanidin biosynthesis in Arabidopsis thaliana. Plant J 39:366–380
Castellarin S, Pfeiffer A, Sivilotti P, Degan M, Peterlunger E, Di Gaspero G (2007) Transcriptional regulation of anthocyanin biosynthesis in ripening fruits of grapevine under seasonal water deficit. Plant Cell Environ 30:1381–1399
Ehlting J, Büttner D, Wang Q, Douglas C, Somssich I, Kombrink E (1999) Three 4-coumarate: coenzyme A ligases in Arabidopsis thaliana represent two evolutionarily divergent classes in angiosperms. Plant J 19:9–20
Eungwanichayapant P, Popluechai S (2009) Accumulation of catechins in tea in relation to accumulation of mRNA from genes involved in catechin biosynthesis. Plant Physiol Biochem 47:94–97
Fang Z, Zhou D, Ye X, Jiang C, Pan S (2016) Identification of candidate anthocyanin-related genes by transcriptomic analysis of ‘Furongli’ plum (Prunus salicina Lindl.) during fruit ripening using RNA-seq. Front Plant Sci 7:1338
Furukawa T, Eshima A, Kouya M, Takio S, Takano H, Ono K (2002) Coordinate expression of genes involved in catechin biosynthesis in Polygonum hydropiper cells. Plant Cell Rep 21:385–389
Gao S (2015) Cloning and functional characterization of a 4-coumarate CoA ligase from liverwort Plagiochasma appendiculatum. Phytochemistry 111:48–58
Ghassempour A, Mollayi S, Farzaneh M, Sharifi-Tehrani A, Aboul-Enein H (2011) Variation of Catechin, epicatechin and their enantiomers concentrations before and after wheat cultivar-Puccinia triticina infection. Food Chem 125:1287–1290
Grzesik M, Naparło K, Bartosz G, Sadowska-Bartosz I (2018) Antioxidant properties of catechins: comparison with other antioxidants. Food Chem 241:480–492
Gui J, Shen J, Li L (2011) Functional characterization of evolutionarily divergent 4-coumarate: coenzyme A ligases in rice. Plant Physiol 157:574–586
Guo F, Guo Y, Wang P, Wang Y, Ni D (2017) Transcriptional profiling of catechins biosynthesis genes during tea plant leaf development. Planta 246:1139–1152
He M, Tian H, Luo X, Qi X, Chen X (2015) Molecular progress in research on fruit astringency. Molecules 20:1434–1451
Hua C, Linling L, Shuiyuan C, Fuliang C, Feng X, Honghui Y, Conghua W (2013) Molecular cloning and characterization of three genes encoding dihydroflavonol-4-reductase from Ginkgo biloba in anthocyanin biosynthetic pathway. PLoS One 8:e72017
Huang J, Gu M, Lai Z, Fan B, Shi K, Zhou Y, Yu J, Chen Z (2010) Functional analysis of the Arabidopsis PAL gene family in plant growth, development, and response to environmental stress. Plant Physiol 153:1526–1538
Huang Y, Gou J, Jia Z, Yang L, Sun Y, Xiao X, Song F, Luo K (2012) Molecular cloning and characterization of two genes encoding dihydroflavonol-4-reductase from Populus trichocarpa. PLoS One 7:e30364
Johnson E, Ryu S, Yi H, Shin B, Cheong H, Choi G (2001) Alteration of a single amino acid changes the substrate specificity of dihydroflavonol 4-reductase. Plant J 25:325–333
Khanizadeh S, Tsao R, Rekika D, Yang R, De Ell J (2007) Phenolic composition and antioxidant activity of selected apple genotypes. J Food Agric Environ 5:61–66
Li P, Du G, Ma F (2011) Phenolics concentration and antioxidant capacity of different fruit tissues of astringent versus non-astringent persimmons. Sci Hortic 129:710–714
Lindermayr C, Möllers B, Fliegmann J, Uhlmann A, Lottspeich F, Meimberg H, Ebel J (2002) Divergent members of a soybean (Glycine max L.) 4-coumarate:coenzyme A ligase gene family. Eur J Biochem 269:1304–1315
Liu M, Tian H, Wu J, Cang R, Wang R, Qi X, Xu Q, Chen X (2015) Relationship between gene expression and the accumulation of catechin during spring and autumn in tea plants (Camellia sinensis L.). Hortic Res 2:15011
Mayr U, Michalek S, Treutter D, Feucht W (1997) Phenolic compounds of apple and their relationship to scab resistance. J Phytopathol-Phytopathol Z 145:69–75
Mosel H, Herrmann K (1974) Changes in catechins and hydroxycinnamic acid derivatives during development of apples and pears. J Sci Food Agric 25:251–256
Nishihara M, Nakatsuka T, Yamamura S (2005) Flavonoid components and flower color change in transgenic tobacco plants by suppression of chalcone isomerase gene. FEBS Lett 579:6074–6078
Pang Y, Peel G, Wright E, Wang Z, Dixon R (2007) Early steps in proanthocyanidin biosynthesis in the model legume Medicago truncatula. Plant Physiol 145:601–615
Rani A, Singh K, Ahuja P, Kumar S (2012) Molecular regulation of catechins biosynthesis in tea [Camellia sinensis (L.) O. Kuntze]. Gene 495:205–210
Samanta T, Kotamreddy J, Ghosh B, Mitra A (2017) Changes in targeted metabolites, enzyme activities and transcripts at different developmental stages of tea leaves: a study for understanding the biochemical basis of tea shoot plucking. Acta Physiol Plant 39:11
Severo J, Tiecher A, Chaves F, Silva J, Rombaldi C (2011) Gene transcript accumulation associated with physiological and chemical changes during developmental stages of strawberry cv. Camarosa. Food Chem 126(3):995–1000
Singh K, Kumar S, Rani A, Gulati A, Ahuja P (2009) Phenylalanine ammonia-lyase (PAL) and cinnamate 4-hydroxylase (C4H) and catechins (flavan-3-ols) accumulation in tea. Funct Integr Genomic 9:125
Soares S, Ferrer-Galego R, Brandão E, Silva M, Mateus N, Freitas V (2016) Contribution of human oral cells to astringency by binding salivary protein/tannin complexes. J Agric Food Chem 64:7823–7828
Tian H (2015) The relationship between cucumber astrengency form and catechins metabolism, and the study of molecular basis. Master’s degree dissertation in Yangzhou University 20–24
Trapnell C, Williams B, Pertea G, Mortazavi A, Kwan G, Van Baren M, Salzberg S, Wold B, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotech 28:511–515
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley D, Pimentel H, Salzberg S, Rinn J, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc 7:562–578
Troszyńska A, Estrella I, Lamparski G, Hernández T, Amarowicz R, Pegg R (2011) Relationship between the sensory quality of lentil (Lens culinaris) sprouts and their phenolic constituents. Food Res Int 44:3195–3201
Xiong L, Li J, Li Y, Yuan L, Liu S, Huang J, Liu Z (2013) Dynamic changes in catechin levels and catechin biosynthesis-related gene expression in albino tea plants (Camellia sinensis L.). Plant Physiol Bioch 71:132–143
Xu W, Lepiniec L, Dubos C (2014) New insights toward the transcriptional engineering of proanthocyanidin biosynthesis. Plant Signal Behav 9:e28736
Xu X, Xu R, Zhu B, Yu T, Qu W, Lu L, Xu Q, Qi X, Chen X (2015) A high-density genetic map of cucumber derived from Specific Length Amplified Fragment sequencing (SLAF-seq). Front Plant Sci 5:768
Xu Y, Zhang Y, Chen J, Wang F, Du Q, Yin J (2018) Quantitative analyses of the bitterness and astringency of catechins from green tea. Food Chem 258:16–24
Yamamoto M, Nakatsuka S, Otani H, Kohmoto K, Nishimura S (2000) (+)-Catechin acts as an infection-inhibiting factor in strawberry leaf. Phytopathology 90:595–600
Zhang L, Wei K, Cheng H, Wang L, Zhang C (2016) Accumulation of catechins and expression of catechin synthetic genes in Camellia sinensis at different developmental stages. Bot Stud 57:31
Zhang Y, Wei K, Li H, Wang L, Ruan L, Pang D, Cheng H (2018) Identification of key genes involved in catechin metabolism in tea seedlings based on transcriptomic and HPLC analysis. Plant Physiol Biochem 133:107–115
Acknowledgements
This work was supported by the National Basic Research Program of China (973 Program; no. 2012CB113900), the Research Innovation Program for College Graduates of Jiangsu Province (no. KYLX15_1374) and the Jiangsu Science and Technology Project (BE2012326). We thank Lesley Benyon, PhD, from Liwen Bianji, Edanz Group China (www.liwenbianji.cn/ac), for editing the English text of a draft of this manuscript.
Author information
Authors and Affiliations
Contributions
XC designed the study; XX, MH and HT carried out the experiments; XX, JP, QX and XQ analyzed the date; XX, JP and MH wrote the paper. All of the authors reviewed and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest in the publication.
Electronic supplementary material
Below is the link to the electronic supplementary material.
13205_2019_1922_MOESM1_ESM.jpg
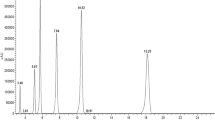
Online Resource 2 (a) HPLC chromatogram of the catechin components in standard solutions. (b) A representative HPLC chromatogram of catechin components in a tested sample (peel of “YanBai” harvested at 6 dpp). (JPEG 194 kb)
Rights and permissions
About this article
Cite this article
Xu, X., Pan, J., He, M. et al. Transcriptome profiling reveals key genes related to astringency during cucumber fruit development. 3 Biotech 9, 390 (2019). https://doi.org/10.1007/s13205-019-1922-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-019-1922-2