Abstract
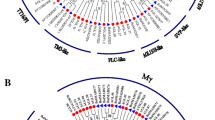
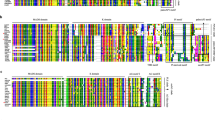
The repertoire and functions of MADS-box family transcription factors (TFs) largely remains unexplored with respect to floral organogenesis of Momordica dioica Roxb. Degenerative PCR followed by rapid amplification of cDNA ends was employed in the present study to clone and characterize 17 MADS-box genes (designated as MdMADS01 to MdMADS17) from the floral buds of M. dioica. The cloned genes were clustered into three subgroups (11 MIKCC, 4 MIKC* and 2 Mα) based on phylogenetic relationships with the MADS-box genes from Cucumis sativus, Cucumis melo and Arabidopsis thaliana. Southern hybridization showed that all the isolated genes were represented by single copy locus in the M. dioica genome. Gene structure analysis revealed 1–8 exons in MdMADS-box genes with the number of exons in MIKC greatly exceeding from that in M-type genes. Motif elicitation of the MdMADS-box genes indicated the presence of additional domains with MIKC type, suggesting that they had more complex structures. Expression analysis of MdMADS genes in six M. dioica transcriptome suggested that, 11 MIKCC—type genes are associated with floral homeotic functions, 4 MIKC*-type genes (MdMADS12 to MdMADS15) controlled the growth of male gametophyte, while the two M-type genes (MdMADS16 and MdMADS17) played significant role in female gametogenesis and seed development. Overall, these are the first set of MADS-box genes from M. dioica exhibiting a differential expression pattern during floral development. The results from this study will provide valuable information for further functional studies of candidate MADS-box genes in the sexual dimorphism of this economically important dioecious cucurbit.





Similar content being viewed by others
References
Adamczyk BJ, Fernandez DE (2009) MIKC* MADS domain heterodimers are required for pollen maturation and tube growth in Arabidopsis. Plant Physiol 149:1713–1723
Bailey TL, Williams N, Misleh C, Li WW (2006) MEME: discovering and analyzing DNA and protein sequence motifs. Nucl Acids Res 34:W369–W373
Baratakke RC, Patil CG (2009) Karyomorphological investigations in dioecious climber Momordica dioica Roxb. J Cytol Genet 11:91–96
Becker A, Theissen G (2003) The major clades of MADS-box genes and their role in the development and evolution of flowering plants. Mol Phylogenet Evol 29:464–489
Bemer M, Wolters-Arts M, Grossniklaus U, Angenent GC (2008) The MADS domain protein DIANA acts together with AGAMOUS-LIKE80 to specify the central cell in Arabidopsis ovules. Plant Cell 20:2088–2101
Bharathi LK, Munshi AD, Vinod Chandrashekaran S, Behera TK, Das AB, John KJ, Nath V (2011) Cytotaxonomical analysis of Momordica L. (Cucurbitaceae) species of Indian occurrence. J Genet 90:21–30
Bhowmick BK, Jha S (2015) Dynamics of sex expression and chromosome diversity in Cucurbitaceae: a story in the making. J Genet 94:793–808
Bhowmick BK, Yamamoto M, Jha S (2016) Chromosomal localization of 45S rDNA, sex specific C values and heterochromatin distribution in Coccinia grandis (L.) Voigt. Protoplasma 253:201–209
Charlesworth D, Mank JE (2010) The birds and the bees and the flowers and the trees: lessons from genetic mapping of sex determination in plants and animals. Genetics 186:9–31
Coen ES, Meyerowitz EM (1991) The war of the whorls: genetic interactions controlling flower development. Nature 353:31–37
Colombo M, Masiero S, Vanzulli S, Lardelli P, Kater MM, Colombo L (2008) AGL23, a type I MADS-box gene that controls female gametophyte and embryo development in Arabidopsis. Plant J 54:1037–1048
De Lucia F, Crevillen P, Am Jones, Greb T, Dean C (2008) A PHD-polycomb repressive complex 2 triggers the epigenetic silencing of FLC during vernalization. Proc Natl Acad Sci USA 105:16831–16836
Diggle PK, Di Stilio VS, Gschwend AR, Golenberg EM, Moore RC, Russell JR, Sinclair JP (2011) Multiple developmental processes underlie sex differentiation in angiosperms. Trends Genet 27:368–376
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:1241–1251
Gramzow L, Theißen G (2010) A hitchhiker’s guide to the MADS world of plants. Genome Biol 11:214
Han P, García-Ponce B, Fonseca-Salazar G, Alvarez-Buylla ER, Yu H (2008) AGAMOUS-LIKE 17, a novel flowering promoter, acts in a FT independent photoperiod pathway. Plant J 55:253–265
Hao X, Fu Y, Zhao W, Liu L, Bade R, Hasi A, Hao J (2016) Genome-wide identification and analysis of the MADS-box gene family in melon. J Am Soc Hort Sci 141:507–519
Heijmans K, Morel P, Vandenbussche M (2012) MADS-box genes and floral development: the dark side. J Expt Bot 63:5397–5454
Hossain MA, Islam M, Ali M (1996) Sexual crossing between two genetically female plants and sex genetics of kakrol (Momordica dioica Roxb.). Euphytica 90:121–125
Hu L, Liu S (2012) Genome-wide analysis of the MADS-box gene family in cucumber. Genome 55:245–256
Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN (2008) The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. Plant Cell 20:635–647
Lee S, Woo YM, Ryu S, Shin YD, Kim WT, Park KY, Lee IJ, An G (2008) Further characterization of a rice AGL12 group MADS-box gene, OsMADS26. Plant Physiol 147:156–168
Litt A, Irish V (2003) Duplication and diversification in the APETALA/FRUITFUL floral homeotic gene lineage: implications for the evolution of floral development. Genetics 165:821–833
Liu C, Chen H, Er HL, Soo HM, Kumar PP, Han JH, Liou YC, Yu H (2008) Direct interaction of AGL24 and SOC1 integrates flowering signals in Arabidopsis. Development 135:1481–1491
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C (T)) method. Methods 25:402–408
Masiero S, Colombo L, Grini PE, Schnittger A, Kater MM (2011) The emerging importance of type I MADS box transcription factors for plant reproduction. Plant Cell 23:865–872
Matias-Hernandez L, Battaglia R, Galbiati F, Rubes M, Eichenberger C, Grossniklaus U, Kater MM, Colombo L (2010) VERDANDI is a direct target of the MADS domain ovule identity complex and affects embryo sac differentiation in Arabidopsis. Plant Cell 22:1702–1715
Melzer S, Lens F, Gennen J, Vanneste S, Rohde A, Beeckman T (2008) Flowering-time genes modulate meristem determinacy and growth form in Arabidopsis thaliana. Nat Genet 40:1489–1492
Ming R, Bendahmane A, Renner SS (2011) Sex chromosomes in land plants. Annu Rev Plant Biol 62:485–514
Mohanty JN, Nayak S, Jha S, Joshi RK (2016) A sequence tagged site (STS) marker encoding Copia-like retrotransposable element is associated with male specific sex expression in Momordiac dioica Roxb. Sci Hort 201:265–270
Mohanty JN, Nayak S, Jha S, Joshi RK (2017) Transcriptome profiling of the floral buds and discovery of genes related to sex-differentiation in the dioecious cucurbit Coccinia grandis (L.) Voigt. Gene 626:395–406
Nesi N, Debeaujon I, Jond C, Stewart AJ, Jenkins GI, Caboche M, Lepiniec L (2002) The TRANSPARENT TESTA16 locus encodes the ARABIDOPSIS BSISTER MADS domain protein and is required for proper development and pigmentation of the seed coat. Plant Cell 14:2463–2479
Parenicová L, de Folter S, Kieffer M, Horner DS, Favalli C, Busscher J, Cook HE, Ingram RM, Kater MM, Davies B, Angenent GC, Colombo L (2003) Molecular and phylogenetic analyses of the complete MADS-box transcription factor family in Arabidopsis: new openings to the MADS world. Plant Cell 15:1538–1551
Pelaz S, Ditta GS, Baumann E, Wisman E, Yanofsky MF (2000) B and C floral organ identity functions require SEPALLATA MADS-box genes. Nature 405:200–203
Renner SS, Ricklefs RE (1995) Dioecy and its correlations in the flowering plants. Am J Bot 82:596–606
Rijpkema AS, Zethof J, Gerats T, Vandenbussche M (2009) The petunia AGL6 gene has a SEPALLATA-like function in floral patterning. Plant J 60:1–9
Schaefer H, Renner SS (2010) A three genome phylogeny of Momordica (Cucurbitaceae) suggests seven returns from dioecy to monoecy and recent long-distance dispersal to Asia. Mol Phylogenet Evol 54:553–560
Shu Y, Yu D, Wang D, Guo D, Guo C (2013) Genome-wide survey and expression analysis of the MADS-box gene family in soybean. Mol Biol Rep 40:3901–3911
Smaczniak C, Immink RGH, Angenent GC, Kaufmann K (2012) Developmental and evolutionary diversity of plant MADS-box domain factors: insights from recent studies. Development 139:3081–3098
Tamura K, Stecher G, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tao Z, Shen L, Liu C, Liu L, Yan Y, Yu H (2012) Genome-wide identification of SOC1 and SVP targets during the floral transition in Arabidopsis. Plant J 70:549–561
Tapia-López R, García-Ponce B, Dubrovsky JG, Garay-Arroyo A, Pérez- Ruíz RV, Kim SH, Acevedo F, Pelaz S, Alvarez-Buylla ER (2008) An AGAMOUS-related MADS-box gene, XAL1 (AGL12), regulates root meristem cell proliferation and flowering transition in Arabidopsis. Plant Physiol 146:1182–1192
Theiben G, Melzer R, Rumpler F (2016) MADS-domain transcription factors and the floral quartet model of flower development: linking plant development and evolution. Development 143:3259–3271
Theißen G (2001) Development of floral organ identity: stories from the MADS house. Curr Opin Plant Biol 4:75–85
Theißen G, Gramzow L (2016) Structure and evolution of plant MADS domain transcription factors. In: Gonzalez DH (ed) Plant transcription factors: evolutionary, structural and functional aspects. Elsevier, Philadelphia, pp 127–138
Tian Y, Dong Q, Ji Z, Chi F, Cong P, Zhou Z (2015) Genome-wide identification and analysis of the MADS-box gene family in apple. Gene 555:277–290
Verelst W, Saedler H, Münster T (2007a) MIKC* MADS-protein complexes bind motifs enriched in the proximal region of late pollen-specific Arabidopsis promoters. Plant Physiol 143:447–460
Verelst W, Twell D, de Folter S, Immink R, Saedler H, Münster T (2007b) MADS-complexes regulate transcriptome dynamics during pollen maturation. Genome Biol 8:R249
Volz SM, Renner SS (2008) Hybridization, polyploidy and evolutionary transitions between monoecy and dioecy in Bryonia (Cucurbitaceae). Am J Bot 95:1297–1306
Vyskot B, Hobza R (2004) Gender in plants: sex chromosomes are emerging from the fog. Trends Genet 20:432–438
Wuest SE, O’Maoileidigh DS, Rae L, Kwasniewska K, Raganelli A, Hanczaryk K, Lohan AJ, Loftus B, Graciet E, Wellmer F (2012) Molecular basis for the specification of floral organs by APETALA3 and PISTILLATA. Proc Natl Acad Sci USA 109:13452–13457
Acknowledgement
The study is funded by a research grant (BT/PR3919/PBD/16/959/2011) from the Department of Biotechnology, Government of India. JNM is grateful to DBT, India for financial support in the form of Senior Research Fellowship. The authors are thankful to Prof. Manoj Ranjan Nayak, President, Siksha O Anusandhan University for his guidance and support. We also thank Prof. Sumita Jha, Centre for Advanced Study, Dept. of Botany, Calcutta University and Prof. Sanghamitra Nayak, Head, Centre of Biotechnology, Siksha O Anusandhan University for their able guidance and support. We also thank DST-FIST, Govt. of India, for the research infrastructure facilities provided to Centre of Biotechnology, Siksha O Anusandhan University.
Author information
Authors and Affiliations
Contributions
RKJ conceived and supervised the project. JNM collected samples, isolated RNA, performed cloning, sequencing and qPCR expression analyses. JNM and RKJ interpreted the data and prepared the manuscript. Both authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors have declared that no competing interest exists.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mohanty, J.N., Joshi, R.K. Molecular cloning, characterization and expression analysis of MADS-box genes associated with reproductive development in Momordica dioica Roxb.. 3 Biotech 8, 150 (2018). https://doi.org/10.1007/s13205-018-1176-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-018-1176-4