Abstract
Bacillus thuringiensis is a major source of insecticidal genes imparting insect resistance in transgenic plants. Level of expression of transgenes in transgenic plants is important to achieve desirable level of resistance against target insects. In order to achieve desirable level of expression, rice chloroplast transit peptide sequence was fused with synthetic cry2AX1 gene to target its protein in chloroplasts. Sixteen PCR positive lines of rice were generated by Agrobacterium mediated transformation using immature embryos. Southern blot hybridization analysis of T0 transgenic plants confirmed the integration of cry2AX1 gene in two to five locations of rice genome and ELISA demonstrated its expression. Concentration of Cry2AX1 in transgenic rice events ranged 5.0–120 ng/g of fresh leaf tissue. Insect bioassay of T0 transgenic rice plants against neonate larvae of rice leaffolder showed larval mortality ranging between 20 and 80 % in comparison to control plant. Stable inheritance and expression of cry2AX1 gene was demonstrated in T1 progenies through Southern and ELISA. In T1 progenies, the highest concentration of Cry2AX1 and mortality of rice leaffolder larvae were recorded as 150 ng/g of fresh leaf tissue and 80 %, respectively. The Cry2AX1 expression even at a very low concentration (120–150 ng/g) in transgenic rice plants was found effective against rice leaffolder larvae.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Globally, more than 3 billion people depend on rice (Oryza sativa, L.) as their staple food, and by 2050 about 30 % more rice must be produced to meet the needs of the growing population (Brookes and Barfoot 2003). One of the major constraints in rice production, throughout the rice growing countries of the world, is the menace of insect pests. Among them the lepidopteran insects, such as yellow stem borer (YSB) (Scirphophaga incertulus Walker), striped stem borer (SSB) (Chilo supressalis Walker) and rice leaffolder (RLF) (Cnaphalocrosis medinalis Guenee) are the major ones, causing significant yield losses up to 10 and 30 % on an average, respectively, each year (Krishnaiah and Varma 2012). The uses of chemical insecticides against the boring insects have not found much effective as the insecticides cannot reach them (Deka and Barthakur 2010). Chemical control besides increasing the cost of production also causes deleterious effects to the ecosystem. The development of insect-resistant lines in rice through conventional breeding has been a challenge due to non-availability of resistance source in germplasm collection (Bhattacharya et al. 2006). Thus, an alternative and the most attractive strategy is to employ the tools of genetic engineering to produce insecticidal proteins by introducing heterologus insecticidal genes.
Insecticidal crystal protein gene (cry gene) of Bacillus thuringiensis (Bt) is effective against important crop pests and widely used in plant genetic engineering. Bt mediated insect-resistant crop technology is the most successful application of agricultural biotechnology in today’s agriculture. Several crops such as tomato, cotton, maize and rice have been successfully transformed with different versions of crystal protein gene of Bt (Mandaokar et al. 2000; Sakthi et al. 2015; Jansen et al. 1997; Chen et al. 2005). The first transgenic rice plant with insect-resistant Bt protein (Cry1Ab) was reported by Fujimoto et al. (1993). Thereafter, several rice varieties have been transformed with genes encoding various Cry1A type Bt crystal proteins and have been shown to be resistant to one or more lepidopteran insect pests of rice (Nayak et al. 1997; Ye et al. 2003; Kim et al. 2009; Wang et al. 2014). First field trial of Bt rice was conducted in China in 1998 (Shu et al. 2000) and in 2009 China’s Ministry of Agriculture issued biosafety certificates for limited commercialization trial in Hubei Province in China for a 5-year period, 2009–2014 (Chen et al. 2011).
The large-scale deployment of transgenic crops expressing a single Bt toxin may lead to break-down of resistance in the field. To delay the resistance breakdown, the concept of gene pyramiding is advocated as one of the resistance management strategies. The cry2A genes are appropriate candidate for pyramiding with cry1A type gene due to variation in their structure and function. Use of synthetic genes expressing hybrid Bt toxins with increased potency is a promising strategy in genetic engineering.
A chimeric Bt gene, cry2AX1 with part of sequence from cry2Aa and cry2Ac was developed in our laboratory to improve the insecticidal activity against a lepidopteran pest, Helicoverpa armigera. The chimeric Cry2AX1 protein exhibited higher level of toxicity than their parental proteins, Cry2Aa and Cry2Ac (Udayasuriyan et al. 2010). The Cry2AX1 protein isolated from recombinant Bt strain was also found to be effective against rice leaffolder. This chimeric gene was codon optimized to improve expression in crop plant (NCBI Accession Number GQ332539.1).
Another concern to be addressed while expressing Bt genes is optimal level of expression for desirable level of protection of crop plants against the target insect pest. One of the promising approaches is to target proteins to specific subcellular sites/compartments of plant cells, such as the chloroplast (Staub et al. 2000). Previously, Jang et al. (1999) reported that the targeting of foreign gene products to plastids using the transit peptide sequence increased the protein product levels in transgenic rice plants.
In the present study, we developed transgenic rice plants with codon optimized synthetic cry2AX1 gene (fused with rice chloroplast transit peptide sequence) using Agrobacterium mediated transformation. The insect bioassay results indicated that the transgenic plants expressing synthetic cry2AX1 gene were resistant to rice leaffolder.
Materials and methods
Construction of plant expression vector and rice transformation
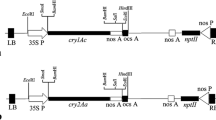
The chimeric cry2AX1 gene was constructed using DNA sequences corresponding to the 585 N-terminal and 48 C-terminal amino acids of Cry2Aa and Cry2Ac, respectively, which were isolated from indigenous strains of Bt. The amino acid sequence of Cry2AX1 differs from that of Cry2Aa for ten residues in a stretch of 24 amino acids (595–618) (Udayasuriyan et al. 2010). Codon optimised, synthetic cry2AX1 gene (Accession Number GQ332539.1) was translationally fused at its 5′ end to the rice chloroplast transit peptide (rtp) sequence of rbcS gene (DNA of 148 bp corresponding to the rice rbcS transit peptide (rtp; Jang et al. 1999). The fusion gene driven by maize ubiquitin constitutive promoter was cloned in pUH binary vector (Agarwal et al. 2002) which harbours hptII (coding for hygromycin phosphotransferase), a plant selectable marker gene. The above construct, pUH-rtp-2AX1 (Fig. 1) was mobilized into disarmed Agrobacterium strain, LBA4404 for rice transformation experiments. A popular rice variety, ASD16 was used for transformation. The Agrobacterium-mediated transformation was carried out according to the protocol described by Hiei and Komari (2008). Regenerated putative transgenic plants were maintained in transgenic greenhouse (Fig. 2).
T-DNA region of plant transformation construct pUH-rtp-2AX1. Rice chloroplast transit peptide (ctp) was fused to cry2AX1 gene. The rtp-cry2AX1 gene is driven by a maize ubiquitin1 promoter and terminated by the nopaline synthase (nos) terminator. The plant selectable marker gene, hptII is under the control of the CaMV35S promoter and tailed by the CaMV35S polyA. LB and RB indicate left border and right border of T-DNA
Agrobacterium-mediated transformation of rice (Oryza sativa L. cv. ASD16). a Immature seeds collected from rice; b pre-treated immature embryos infected with Agrobacterium on cocultivation medium; c callus initiation (and shoot tips) from co-cultivated embryo; d sub-cultured calli on resting medium; e callus proliferation on selection medium; f embryogenic calli on pre-regeneration medium; g regenerated transgenic rice plants; h transgenic rice plants in rooting medium; i established transgenic rice plants in transgenic greenhouse
PCR and Southern blot hybridization analyses
PCR analysis was performed to demonstrate the presence of cry2AX1 and hptII genes in putative transgenic lines of ASD16 using gene specific primers in T0 and T1 transgenic rice plants (Table 1). These primers amplify 800 and 630 bp internal fragments of cry2AX1 and hptII genes, respectively. The plasmid DNA (pUH-rtp-2AX1) was used as positive control and DNA isolated from non-transformed control plants and the reaction mix without template DNA were used as negative control. The amplified PCR products were resolved on 1.2 % agarose gel, visualized on UV transilluminator upon ethidium bromide staining.
Total genomic DNA was extracted from young leaf tissues of transformed plants of both T0 and T1 generation and non-transformed plants using the method described by Dellaporta et al. (1983). For Southern blot analysis, five µg of genomic DNA was digested overnight with HindIII, which has a recognition sequence at one end of the T-DNA. The digested DNA were separated on a 0.8 % agarose gel, and then transferred to a positively charged nylon membrane using 20X saline-sodium citrate (SSC) following standard capillary transfer protocol. The transferred DNA was cross-linked by a UV crosslinker at 1200 μJ min−1 for 1 min. The prehybridisation was carried out for 1 h and hybridization for 18 h at 60 °C. For hybridization, PCR amplified 800 bp internal region of cry2AX1 gene was used as a probe. The probe DNA was labelled with α32P dCTP using Decalabel DNA labelling kit (Thermo Scientific Inc) and added to hybridization solution. After hybridization, the blot was washed with 3X SSC + 0.1 % SDS and 2X SSC + 0.1 % SDS for 15 min each, followed by 10 min in 0.5X SSC + 0.1 % SDS. All washings were carried out at 60 °C and the blot was exposed to X-ray film.
RT-PCR analysis
Total RNA was isolated from PCR positive T1 plants along with a control plant using SV Total RNA Isolation System (Promega, USA) following manufacturer’s instructions. The first strand cDNA was synthesized using RevertAid™ First Strand cDNA Synthesis Kit (MBI Fementas, UK). The primers used for cry2AX1 gene were same as used in PCR assay. The ActF1 and ActR1 primers were used for amplification of actin gene which was used as internal control (Table 1).
Expression of Cry2AX1 protein in transgenic rice plants
The PCR positive T0 transgenic rice plants were subjected for quantitative ELISA at 30 DAS. In T1 progenies the concentration of Cry2AX1 protein was measured at three different phenological stages viz. vegetative (25 DAS), tillering (55 DAS) and reproductive (85 DAS) stages, for studying the temporal variation of cry2AX1 gene expression. About 30 mg of tissues from top of the primary tiller in a plant was used for analysis. The tissue were homogenized in 500 µl of extraction buffer, spun at 3000g at 4 °C for 10 min and 100 µl of the supernatant was immediately used for assay. Each sample was replicated twice. Cry2AX1 protein expression in transgenic rice plants was determined by ELISA kit (Envirologix, USA) following standard procedures. The protein concentration was calculated on a linear standard curve, using the standards provided in the kit.
Detached leaf bit bioassay
Bioassay studies for leaffolder resistance was conducted on ELISA positive T0 and T1 transgenic rice plants to study efficacy of the Cry2AX1 protein. Adults of rice leaffolder were collected from the rice field at Paddy Breeding Station, TNAU, Coimbatore and they were released on susceptible TN1 rice plants maintained in insect cages (65 cm × 65 cm × 75 cm) for culturing. After two generations, the neonate larvae of C. medinalis were used for the bioassay. Leaves of transgenic plants (35 days old seedling) were cut into pieces (about 3 cm length) and three leaf bits were placed on a moist filter paper in a plastic petriplate. Ten neonate larvae of rice leaffolder were released in each petriplate. A control was maintained using leaf bits collected from non-transgenic rice ASD16. Three replications were maintained and the experiment was carried out at 25 °C ± 1, 60 % relative humidity. Larval growth and mortality was recorded every day up to 5 days. After 5 days, leaf area damage and surviving larval characteristics were evaluated in transgenic as well as control plants. Percentage of leaf area damage was calculated using the formula:
Statistical analysis
Values for concentration of Cry2AX1 protein and mortality of rice leaffolder are reported as mean ± SD. The association between protein expression and insect mortality was studied by correlation analysis using the Statistical Package for Social Studies (SPSS) software version 16.
Results
Molecular and biochemical analyses of putative T0 transgenic plants
Twenty putative transgenic rice lines were regenerated and putative transgenic plants were transferred to greenhouse for molecular analyses (Fig. 2). Total genomic DNA from putative rice transformants was subjected to PCR analysis with cry2AX1 and hptII gene specific primers. Out of 20 plants regenerated, 16 were found to be positive for the amplification of ~800 bp (Fig. 3a) and 630 bp internal sequences of cry2AX1 and hptII gene, respectively. Southern hybridization analysis showed two hybridization signals (range 6.7–12.0 kbp) in eight of the 16 plants analyzed whereas the remaining eight plants had three to five signals (range 6.5–19.0 kbp). The non-transformed control plant did not show any hybridization signals (Fig. 3b).
Molecular analysis of T0 Transgenic rice plants. a A 800 bp internal sequence of cry2AX1 gene was amplified by PCR from the DNA isolated from putative transgenic plants. Lanes 1 and 21, 100 bp marker; Lanes 2–17, Putative transgenic plants of ASD16; Lane 18, Non transformed control plant; Lane 19, Negative control (water); Lane 20, pUH-rtp-2AX1 plasmid as a positive control. b Southern blot hybridization analysis of T0 transgenic rice plants expressing cry2AX1 gene. DNA sample isolated from transgenic and non-transgenic plants digested with HindIII restriction enzyme, DNA fragments separated by electrophoresis, transferred to nylon membrane and allowed to hybridize with a radioactively labelled 800 bp internal sequence of cry2AX1 gene. M, λ/HindIII marker; Lanes 1–16, Genomic DNA from transgenic rice plants UR1, UR2, UR3, UR4, UR5 UR6, UR7, UR8, UR9, UR10, UR11, UR12, UR13, UR14, UR15, UR16, respectively; Lane 17, Non-transformed control plant; Lane 18, Positive control (pUH-rtp-2AX1)
The PCR positive transgenic rice lines were further screened by quantitative ELISA kit. All the 16 PCR positive plants were found positive for the expression of Cry2AX1 protein. The expression of Cry2AX1 protein in these transgenic rice lines ranged 10.40–120.50 ng/g fresh leaf tissue (Table 2). One of the T0 plants, UR11 recorded maximum level of expression of Cry2AX1 protein i.e., 120.50 ng/g fresh leaf weight.
Inheritance studies
Thirty plants of T1 progenies derived from transgenic rice line UR11 (selected based on relative levels of Cry2AX1 protein in T0 generation) were established in the greenhouse for further studies on gene inheritance, expression and efficacy. An amplicon of ~800 bp was found in all the 30 T1 progenies for the presence of cry2AX1 gene, whereas no amplification was found in non-transgenic control plant. RT-PCR analysis on a randomly selected five T1 plants showed presence of transcripts of the cry2AX1 (Fig. 4b) and actin gene (Fig. 4a). Further, expression of Cry2AX1 protein in these plants was analyzed at three phenological stages viz, vegetative, tillering and reproductive and the concentration of Cry2AX1 protein varied from 96.00 to 155.00, 46.40 to 86.40 and 30.40 to 72.00 ng/g fresh leaf tissue, respectively (Fig. 4c). The Cry2AX1 protein level in seeds ranged 3.2–4.8 ng/g fresh seed (Supplementary Table 1). Genomic DNA from five ELISA positive T1 progeny plants of UR11 was digested by HindIII enzyme and subjected to Southern blot hybridization using cry2AX1 probe. All the five T1 progenies (UR11–1, UR11–2, UR11–3, UR11–4 and UR11–7) showed three hybridization signals of ~7.5, ~10.0 and ~14.5 kbp (Fig. 5).
a RT-PCR analysis of rice actin (housekeeping gene) in T1 transgenic plants. Lanes 1, 100 bp ladder; Lanes 2–6, transgenic plants (UR11–1, UR11–2, UR11–3, UR11–4 and UR11–7); Lane 7, non-transgenic plants (Wild type ASD16). b RT-PCR analysis of cry2AX1 in T1 transgenic plants. Lane 1 and 9, 100 bp ladder; Lanes 2–6, transgenic plants (UR11–1, UR11–2, UR11–3, UR11–4 and UR11–7); Lane 7, non transgenic plants (Wild type ASD16), Lane 8, Positive control (pUH-rtp-2AX1). c The temporal expression of Cry2AX1 protein in T1 transgenic rice plants. The Cry2AX1 protein concentration in fresh leaf of T1 transgenic plants at vegetative, tillering and reproductive stage
Southern blot hybridization analysis of T1 transgenic rice plants expressing cry2AX1 gene. DNA digested with HindIII and probed with a radioactively labelled 800 bp internal sequence of cry2AX1 gene. M, λ/HindIII marker; Lane 1, UR11–1; Lane 2, UR11–2; Lane 3, UR11–3; Lane 4, UR11–4; Lane 5, UR11–7; Lane 6, Control plant; Lane 7, Positive control (pUH-rtp-2AX1)
Detached leaf bit bioassay
In order to determine the insecticidal activity of Cry2AX1 protein expressed in transgenic rice plants, detached leaf bit bioassay was carried out using neonate larvae of rice leaffolder on the ELISA positive T0 plants. The larval mortality ranged 23.33–80.00 % (Table 2). A significant, positive correlation was found between the concentration of Cry2AX1 and insect mortality in T0 transgenic plants (r = 0.920, p = 0.01). No larval mortality was observed on control plants and the major portion of the leaf tissue was consumed by the surviving larvae over a period of 5 days (Supplementary Figure 1).
The efficiency of T1 transgenic plants (Three T1 progeny plants) expressing Cry2AX1 protein (which expressed relatively higher levels of Cry2AX1 protein) with respective controls were tested against the neonates of rice leaffolder. Mortality in rice leaffolder varied from 53.33 to 80.00 % in the T1 transgenic rice plants (Table 3). The percentage of leaf area damaged by rice leaffolder larvae varied from 20.29 to 36.32 % in the T1 transgenic lines compared to 92.73 % in control plants (Fig. 6).
Discussion
Transgenic plants with insecticidal crystal protein from Bacillus thuringiensis could drastically reduce the use of broad-spectrum insecticides against insect pests. However, there is a risk that insect could become resistant to the Bt toxin. Therefore, it is envisaged that gene pyramiding in transgenic plants could be apossible strategy for simultaneous expression of two or more cry genes (Datta et al. 2002). To improve the insecticidal property of the Cry toxin, a chimeric Bt gene (cry2AX1) was developed with cry2Aa and cry2Ac sequences from indigenous isolates of Bt. Our earlier study proved that the chimeric Cry2AX1 protein expressed in Bt was highly toxic to the larvae of the rice leaffolder, C. medinalis (Manikandan et al. 2014).
A number of studies have suggested that chloroplasts are ideal sites for the expression of transgenes (Jang et al. 1999; Kim et al. 2009; Verma and Daniell 2007). Earlier studies reported improved levels of expression of foreign genes by targeting them to the chloroplast in transgenic tobacco, cotton and rice (Jang et al. 1999; Kim et al. 2009; Rawat et al. 2011). In the present study, we introduced a codon optimised synthetic cry2AX1 gene driven by ubiquitin promoter with rice chloroplast transit peptide for targeting Cry2AX1 protein into the chloroplast.
In this study, twenty putative transformants of rice were generated on hygromycin selection, of which 16 plants were found to be positive for the cry2AX1 gene in PCR assay. The presence of transgene was confirmed by expression analysis of 16 PCR positive T0 transgenic plants by ELISA. All the 16 primary transformants gave positive results for the expression of Cry2AX1 protein and concentration of Cry2AX1 protein in putative transformants ranged 10.40–120.50 ng/g of leaf tissue on fresh weight. Similar variation in expression level was reported by earlier researcher on Bt rice (Wu et al. 2001). Genetic background and gene constructs were shown to influence the level of expression (Maqbool et al. 2001; Breitler et al. 2000; Meiyalaghan et al. 2006). Plant to plant variation in expression is mainly due to integration of transgenic DNA into regions of the genome that are transcriptionally repressed (heterochromatin), which ultimately leads to transgene silencing. These types of position effects often result in the production of transgenic lines exhibiting high and low levels of expression (De Bolle et al. 2003).
Stable integration of transgene was confirmed by Southern hybridization analysis and Southern results indicated integration of cry2AX1 gene at two to five locations in the rice genome. In most of the cases, integration of cry2AX1 gene was at two loci (a relatively simple pattern) and in a few cases, it was 3–5 (complex pattern). Several earlier researchers reported that single copy transgenes had exhibited higher expression levels than multiple T-DNA insertions (Hobbs et al.1990; Jones et al. 1987). However, we observed that level of expression of transgene and resistance was higher in the multiple copy line UR11 compared to other transgenic lines. Zaidi et al. (2009) also reported that the level of Cry1C expression was higher in transgenic rice plant with three copies of cry1C gene when compared to single copy plants.
Detached leaf bit bioassay of the selected T0 plants against neonate larvae of rice leaffolder revealed significant variability in larval mortality. The mortality varied from 23.33 to 80.00 % among the T0 transformants. The effect of the Cry2AX1 protein was also seen on the larva as there was considerable differences in the size of the larva that fed on the transgenic and wild type. Correlation between the Cry2AX1 protein expression and larval mortality showed a highly significant positive relationship (r = 0.920) indicating that higher the concentration of Cry2AX1 protein has resulted in higher mortality of the larvae. Transformants expressing higher levels of Cry2AX1 protein invariably induced higher larval mortality.
The T1 progenies of one of the primary transformants (UR11) were subjected to PCR analysis to determine the stable inheritance of transgene. The Mendalian segregation ratio could not be determined due to smaller size (30) of the T1 progeny. Southern hybridization analysis revealed three integrations of cry2AX1 gene in five of the T1 progenies tested at random (event UR11).
Temporal variation of Cry2AX1 protein was quantified in T1 transgenic rice plants (Five T1 progeny plants) at three phenological stages (vegetative, tillering and reproductive). The results showed a decline in expression of Cry2AX1 when the crop progressed towards the maturity. The level of expression was relatively higher at vegetative stage when compared to other growth periods. One of the progenies i.e., UR11–3 showed the highest level of expression 155.00 ng/g of fresh leaf weight and the transgenic seeds showed significantly low level of expression varying from 3.2 to 4.8 ng/g fresh seed. The ELISA results indicated that the expression of Cry2AX1 protein in leaves decreased towards the end of growing season. Similar results were also reported in previous researches on Bt rice (Wu et al. 2001; Zhao et al. 2004; Han et al. 2009). Wan et al. (2005) reported that expression pattern of Bt toxin in progenies of cotton varies within the season with higher concentration at the beginning and lower at the latter stages. The possible reason for reduction of Bt protein expression is that the level of cry gene transcripts decreased in plant tissues as they mature and is likely because of decreased activity of the promoters, which is probably hindered by adverse physiological conditions in the rice plants (Christensen et al. 1992). Bakhsh et al. (2012) reported that the reduction of Bt protein content in later season of cotton tissue could be attributed to the over expression of the Bt gene at earlier stage, which leads to gene regulation at post-transcriptional level and consequently results in gene silencing at a later stage.
Insect bioassay on T1 transgenic rice plants resulted in larval mortality ranging from 53.33 to 80.00 % with a low level of leaf damage (20.29–36.32 %) whereas in control plant there was no mortality and major portion of leaf tissue (92.73 %) was consumed by surviving larvae over the period of 5 days. The larvae recovered from control plants were well-developed and progressed to next instar (third instar), while the larvae recovered from those of transgenic plants were dead or sluggish, weak and underdeveloped. The reduction in the larval size was evident among the larvae tested on the transgenic rice in the bioassay. This may be because of low level of feeding, which in turn resulted in reduced accumulation of toxins in insects that did not cause the mortality but brought about the reduction in size.
It has been demonstrated by earlier researchers that the Cry protein concentration is directly related to the level of insect-resistance. Chen et al. (2005) reported an insect mortality of 100 % against rice stem borer in plants with average level of Cry2A protein expression of 10 µg/g fresh weight. Riaz et al. (2006) reported a concentration of Cry1Ac protein ranging between 4.6 and 16 µg/g and Cry2A at a level of 0.34–1.45 µg/g of tissue in leaves of transgenic rice plants. In our studies, the generated cry2AX1 transgenic rice plants showed low level of Cry2AX1 expression. Therefore, the level of expression of Cry protein reported by earlier workers indicate that a relatively higher level of cry2AX1 gene expression may be necessary for a desirable level of insecticidal activity in rice plants and this can be achieved by generating and screening more number of putative transformants.
References
Agarwal KS, Kapoor A, Grover A (2002) Binary cloning vectors for efficient genetic transformation of rice. Curr Sci 82:873–876
Bakhsh A, Rao AQ, Shahid AA (2012) Spatio temporal expression pattern of an insecticidal gene (cry2A) in transgenic cotton lines. Not Sci Biol 4:115–119
Bhattacharya J, Mukherjee R, Banga A, Dandapat A, Mandal CC, Hossain MA (2006) A transgenic approach for developing insect resistant rice plant types. Science, Technology and Trade for peace and prosperity (IRRI, ICAR). Mc. Millan India Ltd, New Delhi, pp 245–264
Breitler JC, Marfa V, Royer M, Meynard D, Vassal JM, Vercambre B, Frutos R, Messeguer J, Gabarra R, Guiderdoni E (2000) Expression of a B. thuringiensis cry1B synthetic gene protects Mediterranean rice against the striped stem borer. Plant Cell Rep 19:1195–1202
Brookes P, Barfoot GB (2003) GM rice, will this be the way for global acceptance of GM crop technology. ISAAA Briefs no. 28:ISAAA, Ithaca
Chen H, Tang W, Xu CG, Li XH, Lin YJ, Zhang QF (2005) Transgenic indica rice plants harboring a synthetic cry2A gene of B. thuringiensis exhibit enhanced resistance against rice lepidopteran pests. Theor Appl Genet 111:1330–1337
Chen M, Shelton A, Ye GY (2011) Insect-resistant genetically modified rice in China: from research to commercialization. Ann Rev Entomol 56:81–101
Christensen AH, Sharrock RA, Quail PH (1992) Maize polyubiquitin genes: structure, thermal perturbation of expression and transcript splicing, and promoter activity following transfer to protoplasts by electroporation. Plant Mol Biol 18:675–689
Datta K, Baisakh N, Thet KM, Tu J, Datta SK (2002) Pyramiding transgenes for multiple resistance in rice against bacterial blight, stem borer and sheath blight. Theor Appl Genet 106:1–8
De Bolle MFC, Butaye J, Coucke WJW, Goderis WM, Wouters PFJ, Boxel N, Broekaert WF, Cammue BPA (2003) Analysis of the influence of promoter elements and a matrix attachment region on the inter-individual variation of transgene expression in populations of Arabidopsis thaliana. Plant Sci 165:69–179
Deka S, Barthakur S (2010) Overview on current status of biotechnological interventions on yellow stem borer Scirpophaga incertulas (Lepidoptera: Crambidae) resistance in rice. Biotechnol Adv 28:70–81
Dellaporta SL, Wood J, Hicks JB (1983) A plant DNA minipreparation: version II. Plant Mol Biol Rep 1:873–876
Fujimoto H, Itoh K, Yamamoto M, Kyozuka J, Shimamoto K (1993) Insect resistant rice generated by introduction of a modified gene of B. thuringiensis. Biotechnol 11:1151–1155
Han LZ, Liu PL, Wu KM, Peng YF, Wang F (2009) Population dynamics of Sesamia inferens on transgenic rice expressing Cry1Ac and CpTI in southern China. Environ Entomol 37:1361–1370
Hiei Y, Komari T (2008) Agrobacterium-mediated transformation of rice using immature embryos or calli induced from mature seed. Nat Protocol 3:824–834
Hobbs SLA, Kpodar P, DeLong CMO (1990) The effect of T-DNA copy number, position and methylation on reporter gene expression in tobacco transformants. Plant Mol Biol 15:851–864
Jang IC, Nahm BH, Kim JK (1999) Sub-cellular targeting of green fluorescent protein to plastids in transgenic rice plants provides a high-level expression system. Mol Breed 5:453–461
Jansen SV, Dickburt A, Buysse C, Piens L, Saey C, De Wulf B, Gossele A, Paez VA, Gobel E (1997) Transgenic corn expressing a Cry9C insecticidal protein from Bacillus thuringiensis protected from European corn borer damage. Crop Sci 37:1616–1624
Jones JDG, Gilbert DE, Grady KL, Jorgenesn RA (1987) T-DNA structure and gene expression in petunia plants transformed by Agrobacterium tumefaciens C58 derivatives. Mol Gen Genet 207:478–485
Kim EH, Suh SC, Park BS, Shin KS, Kweon SJ, Park SH, Kim YS, Kim JK (2009) Chloroplast-targeted expression of synthetic cry1Ac in transgenic rice as an alternative strategy for increased pest protection. Planta 230:397–405
Krishnaiah K, Varma NRG (2012) Changing insect pest scenario in the rice ecosystem—a national perspective. Directorate of Rice Research Rajendranagar, Hyderabad. 2012:2–8
Mandaokar AD, Goyalb RK, Shukla A, Bisaria S, Bhalla R, Reddy VS, Chaurasia A, Sharma RP, Altosaar I, Kumar PA (2000) Transgenic tomato plants resistant to fruit borer (Helicoverpa armigera Hubner). Crop Prot 19:307–312
Manikandan R, Balakrishnan N, Balasubramani V, Sudhakar D, Udayasuriyan V (2014) Comparative toxicity of chimeric Cry2AX1 Bt protein isolated from recombinant Bt and E. coli hosts against rice leaf folder (Cnaphalocrosis medinalis). Trends Biosci 7:1125–1130
Maqbool SB, Riazuddin S, Loc NT, Gatehouse AMR, Gatehouse JA, Christou P (2001) Expression of multiple insecticidal genes confers broad resistance against a range of different rice pests. Mol Breed 7:85–93
Meiyalaghan S, Jacobs JME, Butler RC, Wratten SD, Conner AJ (2006) Expression of cry1Ac9 and cry9Aa2 genes under a potato light-inducible Lhca3 promoter in transgenic potatoes for tuber moth resistance. Euphytica 147:297–309
Nayak KP, Basu D, Das S, Basu A, Ghosh D, Ramakrishna NA, Ghosh M, Sen SK (1997) Transgenic elite indica rice plants expressing CryIAc δ-endotoxin of B. thuringiensis are resistant against yellow stemborer (Scirpophaga incertulas). Proc Natl Acad Sci USA 94:2111–2116
Rawat P, Singh AK, Ray K, Chaudhary B, Kumar SV, Gautam T, Kanoria S, Kaur G, Kumar P, Pental D, Burma PK (2011) Detrimental effects of expression of Bt endotoxin Cry1Ac on in vitro regeneration, in vivo growth and development of tobacco and cotton transgenics. J Biosci 36:363–376
Riaz N, Husnain T, Fatima T, Makhdoom R, Bashir K, Masson L, Altosaar I, Riazuddin S (2006) Development of indica rice harboring two insecticidal genes for sustainable resistance against lepidopteran insects. S Afr J Bot 72:217–223
Sakthi AR, Naveenkumar A, Deepikha PS, Balakrishnan N, Kumar KK, Kokila Devi E, Balasubramani V, Arul L, Singh PK, Sudhakar D, Udayasuriyan V, Balasubramanian P (2015) Expression and inheritance of chimeric cry2AX1 gene in transgenic cotton plants generated through somatic embryogenesis. In Vitro Cell Dev Biol Plant. doi:10.1007/s11627-015-9695-8
Shu Q, Ye G, Cui H, Cheng X, Xiang Y, Wu D, Goa M, Xia Y, Hu C, Sardana R, Altosaar I (2000) Transgenic rice plants with a synthetic cry1Ab gene from B. thuringiensis were highly resistant to eight Lepidopteran pest species. Transgenic Res 9:433–439
Staub JM, Garcia B, Graves J, Hajdukiewicz PTJ, Hunter P, Nehra N, Paradkar V, Schlittler MJ, Carroll A, Ward D (2000) High-yield production of a human therapeutic protein in tobacco chloroplasts. Nat Biotechnol 18:333–338
Udayasuriyan V, Indra Arulselvi P, Balasubramani V, Sudha DR, Balasubramanian P, Sangeetha P (2010) Construction of new chimeric cry2AX1 gene of B. thuringiensis encoding protein with enhanced insecticidal activity. Indian Patent number 244427
Verma D, Daniell H (2007) Chloroplast vector systems for biotechnology applications. Plant Physiol 145:1129–1143
Wan P, Zhang Y, Wu K, Huang M (2005) Seasonal expression profiles of insecticidal protein and control efficacy against Helicoverpa armigera for Bt cotton in China. J Econ Entomol 98:195–201
Wang Y, Zhang L, Li Y, Liu Y, Han L, Zhu Z, Wang F, Peng Y (2014) Expression of Cry1Ab protein in a marker-free transgenic Bt rice line and its efficacy in controlling a target pest, Chilo suppressalis (Lepidoptera: Crambidae). Environ Entomol 43:528–536
Wu G, Cui HR, Shu QY, Ye GY, Xie XB, Xia YW, Gao MW, Altosaar I (2001) Expression patterns of cry1Ab gene in progenies Kemingdao and the resistance to striped stem borer. Sci Agric Sin 34:465–468
Ye GY, Yao HW, Shu QY (2003) High levels of stable resistance in transgenic rice with a cry1Ab gene from B. thuringiensis Berliner to rice leaffolder, Cnaphalocrocis medinalis (Guenee) under field conditions. Crop Prot 22:171–178
Zaidi MA, Ye GY, Yao HW (2009) Transgenic rice plants expressing a modifed cry1Ca1 gene are resistant to Spodoptera litura and Chilo suppressalis. Mol Biotechnol 43:232–242
Zhao HY, Zhang YJ, Wu KM, Zhao KJ, Peng YF, Guo YY (2004) Expression of CrylAc protein in Cry1Ac/CpTI transgenic rice and its resistance in different stages to Chilo suppressalis. J Agric Biotechnol 12:76–79
Acknowledgments
We thank Prof. Dr. S. Suresh, Dr. R. P. Soundararajan, Assistant professor (Department of Entomology, TNAU, Coimbatore) for providing the facilities for rearing of rice leaffolder. We thank Dr. K. K. Kumar (Department of Plant Biotechnology, TNAU) for his help in Southern analysis.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors certify that there is no conflict of interest in our manuscript.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Manikandan, R., Balakrishnan, N., Sudhakar, D. et al. Transgenic rice plants expressing synthetic cry2AX1 gene exhibits resistance to rice leaffolder (Cnaphalocrosis medinalis). 3 Biotech 6, 10 (2016). https://doi.org/10.1007/s13205-015-0315-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-015-0315-4