Abstract
Phosphoenolpyruvate carboxykinase (PCK) catalyzes the decarboxylation and phosphorylation of oxaloacetate to phosphoenolpyruvate in the gluconeogenic pathway in most organisms. A pckA gene encoding PCK was cloned and sequenced from strain Rhizobium pusense NRCPB100, a spontaneous rifampicin resistant mutant of R. pusense NRCPB10T (JCM16209T) of a recently described new species of a non-nodulating and non-tumorigenic bacterium. The mapping of the pck gene region following Tn5 mutagenesis located the gene downstream of a transcriptional regulatory protein gene (chvI) and upstream of a conserved hypothetical protein gene. The pck of 1,611 bp was deduced to encode 536 amino acids and showed high homology to the genes of known ATP-dependent PCK enzymes. Phylogenetic analysis of the gene placed it in a cluster with pck of other known members of Rhizobiales. Amino acid sequences of the putative functional regions of the deduced enzyme were found to be conserved.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Gluconeogenesis refers to the central metabolic pathway in which tricarboxylic acid (TCA) cycle intermediates such as oxaloacetate and malate are converted to hexose sugars to maintain hexose sugar homeostasis in cells. In many organisms, the first step of the gluconeogenic pathway is catalyzed by the enzyme phosphoenolpyruvate carboxykinase (PCK) (EC 4.1.1.49) which decarboxylates and simultaneously phosphorylates oxaloacetate to phosphoenolpyruvate (PEP) (Utter and Kolenbrander 1972). Fructose-1,6-bisphosphatase, which converts fructose-1,6-bisphosphate to fructose-6-phosphate, is required for gluconeogenesis (Østerås et al. 1997; Voet et al. 2008). PCK has also been demonstrated to play a crucial role in several metabolic processes associated with cataplerosis (Yang et al. 2009). PCKs have been classified according to their specificities for nucleotides, GTP and ATP. Generally, the enzymes from mammals and a number of other eukaryotes have specificity for GTP, while the enzymes from bacteria, yeast and plants have specificity for ATP (Utter and Kolenbrander 1972). In addition, all eukaryotes studied to date, also have copies of mitochondrial PCK (PCK-M) and cytosolic PCK (PCK-C) (Yang et al. 2009). PCK also appears to have an absolute requirement for divalent metal ions, such as Mn2+, but other divalent ions, such as Mg2+ or Co2+, can act as substitutes but with reduced activity (Utter and Kolenbrander 1972).
Besides the PCK reaction an alternative pathway exists in bacteria e.g. in Escherichia coli, in which oxaloacetate or malate is decarboxylated to pyruvate by oxaloacetate decarboxylase or malic enzyme, respectively (Hansen and Juni 1974). The pyruvate formed is then converted to PEP by PEP synthetase or by pyruvate: orthophosphate dikinase (Cooper and Kornberg 1967). Strains deficient in both oxaloacetate decarboxylase and malic enzyme cannot grow on dicarboxylic acid intermediates of the TCA cycle as sole carbon source.
Free-living Rhizobium leguminosarum bv. viciae (formerly Rhizobium leguminosarum) and Ensifer meliloti (formerly R. meliloti, then Sinorhizobiummeliloti) and Rhizobium sp. strain NGR234 require PCK, fructose-1,6-bisphosphate aldolase and fructose-1,6-bisphosphatase for growth and metabolism of C4-dicarboxylates through the gluconeogenic pathway (McKay et al. 1985, 1988; Finan et al. 1988; Østerås et al.1991). PCK deficient mutants of Rhizobium species fail to grow on minimal medium with succinate or other TCA cycle intermediates but are able to grow on glucose or glycerol as the sole carbon source suggesting that PCK functions in Rhizobium as a key gluconeogenic enzyme (Østerås et al. 1991). However, in E. melilotipckA appears to be regulated differentially in the bacteroids than in free-living cells. During symbiosis, the host plant supplies C4-dicarboxylates, succinate and malate, to the bacteroids within nodules. The low PCK activities in the symbiotic phenotypes of PCK mutants of E. meliloti suggest that the PCK pathway is not operative in N2-fixing bacteroids and that PCK may be important in the process of infection rather than in gluconeogenesis. However, the understanding of the role of pckA in the root nodule symbiosis is further complicated by the finding that RhizobiumpckA mutants are symbiotically defective in some host plants but are phenotypically a wild type on other hosts (Østerås et al. 1991). In E. coli, pckA expression is induced by gluconeogenic substrates and the expression is regulated by the growth phase of the culture and by cAMP (Goldie 1984). In Agrobacterium pckA is induced by acidic pH and is involved in the expression of vir genes (Liu et al. 2005). PCK is essential for growth of Mycobacterium tuberculosis on fatty acids and the organism relies on gluconeogenesis to establish and maintain infection (Merrero et al. 2010). Deletion of the pckA in M. bovis led to the reduction in the capacity of the bacterium to infect and survive in macrophages suggesting that PCK activity is important during establishment of infection (Liu et al. 2003). On the other hand, PCK-C mRNAs in human were found to be more abundant in non-tumor tissues than in the tumors invoking the gene for PCK-C to define as a new negative marker for colonic tumors (Bluin et al. 2010). It appears that analysis of pckA and gene products may yield new information on the requirements for infection and for survival of a non-infective species.
Recently, we have isolated R. pusense NRCPB10T(JCM16209T = LMG25623T = NCIMB 14639T) a new species from the rhizospheric soil of chickpea (Cicer arietinum L), a close phylogenetic relative of R. radiobacter (Panday et al. 2011), which was previously known as A. tumefaciens (Young et al. 2001). The present investigation was carried out in search for a possible reason for its non-infectivity. Previously PCK− mutants of R. leguminosarum bv. viciae were reported to have no symbiotic phenotype (Finan et al. 1988) and pckA mutant of A. tumefaciens was also shown to be highly attenuated in tumor inducing ability. These observations prompted us to study the pckA gene of R. pusense NRCPB10 for its non-infectivity. Here we report the cloning, physical localization, nucleotide sequence and phylogeny of the pckA gene of R. pusense NRCPB100.R. pusense NRCPB100 is a spontaneous rifampicin resistant mutant of the strain NRCPB10.
Materials and methods
Bacterial strains, plasmids and media
The bacterial strains and plasmids used in this study are listed in Table 1. R. pusense NRCPB10 was grown on yeast mannitol (YM) agar medium (Vincent 1970) containing rifampicin (50 μg/ml) and a rifampicin resistant spontaneous mutant strain NRCB100 was isolated. NRCB100 was grown either in YM medium or in rhizobium minimal medium (RMM), pH 6.8, containing: (g/l): K2HPO4, 2; KH2PO4, 1.5; NaCl, 0.15; NH4Cl, 0.5; MgSO4·7H2O, 0.5; CaCl2.2H2O, 0.01 and glucose, 10 as the carbon source.E. coli was grown in Luria broth (Miller 1972). The media were solidified with agar powder (Difco, USA) at 15 g/l when necessary.
Tn5 mutagenesis
R. pusense NRCPB100 grown in YM for 2 days at 28 °C was mutagenised by mating with the donor E. coli WA803 (Wood 1996) harboring the suicide plasmid pGS9:: Tn5 (Selvaraj and Iyer 1983) grown in Luria broth for 12 h at 37 °C as described (Gautam et al. 2007). Transconjugants were selected by plating dilutions of the bacteria on RMM containing glucose plus succinate as the carbon sources and supplemented with 2,3,5-triphenyl tetrazolium chloride (30 μg/ml), rifampicin (50 μg/ml) and neomycin (100 μg/ml). After incubating the plates for 3 days white colonies of mutants defective in the utilization of dicarboxylic acids were isolated (Ronson et al. 1981). The mutants failed to grow on succinate as the carbon source.
Bacterial growth
Growth of R. pusense NRCPB100 and the mutant NRC43 was analyzed in RMM containing succinate (10 g/l) as the carbon source. Inocula were prepared by growing the bacteria in YM medium and washing with RMM. Growth of the bacteria in RMM was monitored by measuring the optical density at 600 nm in a SPECORD210 spectrophotometer, Analytik Jena, Germany.
DNA manipulation, cloning and sequencing
DNA was isolated according to standard protocol (Meade et al. 1982). Routine manipulation of DNA, plasmid isolation, construction of recombinant plasmids, electrophoresis of DNA and transformation were carried out according to standard procedures (Sambrook and Russell, 2001). Digestions with restriction enzymes and DNA ligation were performed according to manufacturers’ instructions (Promega, Inc. and New England Biochemical, Beverly, Mass.). Southern blotting was performed as described (Southern 1975).
Sequencing of the EcoRI DNA fragment of NRC43 containing Tn5, which was cloned in pBlueScript vector, was performed with T3 and T7 primers by a commercial source (Microsynth GmbH, Switzerland) using an Applied Biosystems DNA Sequencer and a ABI PRISM Big Dye Terminator Cycle Sequencing kit (Applied Biosystems). Reaction products were analyzed with ABI PRISM 310 Genetic Analyzer (Applied Biosystem). After the first round, based on the partial sequence worked out new internal forward and reverse primers were designed and used to proceed with the next round of sequencing and the process was repeated each time with newly designed primers. Finally the chains of nucleotide sequences were assembled into a contiguous 2.737 kb sequence. ORFs were identified using the ORF finder facility of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/gorf/gorf.html).
The oligonucleotide primers F1 (5′-ATGACCGAGATTGGGGTTCATAAT-3′) and R1 (5′-TTCTGCCGCGAGCAAAAGGCCCGG-3′) were designed from the sequences of the flanking regions of pckA gene of NRC 43 and were used for PCR amplification of 1.611 kb stretch of DNA, the complete coding region of pckA gene (ORF2), using R. pusense NRCPB100 genomic DNA as template. The resulting amplicon was purified and cloned into pGEM-TEasy plasmid vector (Promega, USA) for sequencing.
Construction of plasmids for complementation
The oligonucleotide primers pckAF (5′-GCCGAATTCATTGTGGCAAAGGAAAAC-3′) and pckAR (5′-GATGAATTCTTCTGCCGCGAGCAAAAG-3′), designed from the DNA sequence of NRC 43 and were used for PCR amplification of about 1.851 kb of genomic DNA, which contained the pckA gene (ORF2) along with its promoter sequence, introducing the EcoRI site (underlined) into the PCR product. After digestion with EcoRI the amplicon was cloned into pSUP104 (Figurski and Helinski 1979) and E. coli HB101 was transformed with it to yield kSD3. The plasmid kSD3 was used to complement NRC43. Triparental spot mating was performed using the recipient NRC43, the donor kSD3 and the helper strain HB101/pRK2013 (Priefer et al. 1985). The ex-conjugants were identified by streaking on RMM containing glucose as the carbon source and supplemented with tetracycline and rifampicin.
Genomic context analysis
Genomic context of pckA of R. pusense NCRPB10 in comparison with the genomes of related members of Rhizobiales was analysed using the GeConT programme. This programme is available at: http://www.ibt.unam.mx/biocomputo/gecont.html (Ciria et al. 2004).
Phylogenetic analysis of the pckA gene
The EcoRI fragment of NRC43 containing Tn5 was cloned in pBlueScript, sequenced, and a chain of 2.757 kb of nucleotides, which contained the pckA gene, was assembled. The ORFs in the sequence identified using BLASTX. BLAST searches provided the homologous sequences of the gene from the databases (Altschul et al. 1997). A neighbour-joining phylogenetic tree was constructed using pckA gene sequences of different organisms as per Kimura’s 2 parameter model (Kimura 1980) using MEGA software package, version 4.0 (Tamura et al. 2007). The tree topologies and statistical significance of branch points of the distance were tested by 1,000 bootstrap re-samplings of the data (Felsenstein 1985). The deduced amino acid sequences of the pckA genes were aligned using the ClustalW program package from EMBL (Heidelberg, Germany).
Nucleotide sequence accession number
The nucleotide sequence data of pckA of R. pusense were deposited in GenBank database under accession number AF450091.
Results and discussion
Transposon mutagenesis and isolation of mutant
Tn5 was introduced into R. pusense strain NRCPB100 using the suicide plasmid pGS9 containing Tn5 from E. coli WA803 (Wood 1996). Transconjugants occurred at a frequency of 3.2 × 10−4 per donor. Transconjugants were selected on YM-agar plates containing neomycin and rifampicin. Over five thousand Neor, Rifr transconjugants were obtained. Screening on RMM containing glucose plus succinate as the carbon sources and 2,3,5-triphenyl tetrazolium chloride allowed the isolation of several white mutants of which NRC43 (Table 1) was arbitrarily chosen for further studies.
The mutant NRC43 failed to grow in RMM containing the glucogenic substrate succinate as was also shown for pckA mutants of E. meliloti, R. leguminosarum bv. viciae and Rhizobium sp. NGR234 (Finan et al. 1988; Østerås et al. 1991, 1995), whereas growth of the strain on mannitol or glucose as carbon source was similar to that of its parent. NRC43 also had growth, similar to its parent, on glucose, fructose, galactose, mannose, rhamnose, arabinose, xylose, lactose, maltose, mannitol or sorbitol as the carbon source but not on succinate, malate, fumarate, or pyruvate. The data indicated that the mutation in NRC43 did not allow it to grow in glucogenic substrates as the sole carbon and energy source, presumably the strain lacked the key enzyme of gluconeogenesis. On the other hand, growth of the parent NRCPB100 in the glucogenic substrates indicated that glucogenesis was operative in the strain, presumably due to induction of the key enzyme in the gluconeogenic pathway.
Cloning and physical localization of the gene complementing NRC43
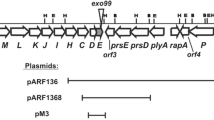
The EcoRI DNA fragment from R. pusense NRC43 containing Tn5 was cloned into the pBluescript vector, transformed into E. coli strain DH5α and the recombinant plasmid kSD1 was isolated. The nucleotide sequence of the DNA flanking Tn5 in kSD1 was determined using T3 and T7 primers. The sequence revealed one complete and two partial open reading frames (ORFs) (Fig. 1). Database sequence similarity searches showed that in the strain NRC43, Tn5 had interrupted the gene homologous to pckA gene. The gene was flanked by two open reading frames; ORF1 upstream and ORF3 downstream of it. Analysis of the sequence of the 2.737 kb fragment also revealed that the ORF1 had the ATG start codon at position 851 which was preceded by a potential ribosome binding site, AGGAA, located 11 bp upstream. The putative transcription start site, AAAT, at position 730 was preceded by potential −10 (AAATAAAAT) and −35 (TTCAAC) sequences at positions 716 and 698 respectively, which were separated by 18 bp.
Genetic organization and genomic context of Rhizobium pusense NRC43 pckA DNA region. a Genetic organization of the pckA DNA region of R. pusense NRC43 and the transcriptional directions of ORF1, ORF2 (pckA) and ORF3 in the EcoRI digested DNA fragment containing Tn5. b Conservation of the genomic context of the pckA region in the genome of R. pusense in comparison with that of related members of Rhizobiales. Arrows represent genes with their relative orientation in the genomes. Genes are colored according to their functional categories and labeled according to their original gene annotation in the database. Yellow, two component response regulator; orange, phosphoenol pyruvate carboxykinase; red, hypothetical protein; blue, pantothenate kinase; black, alkylated DNA repair protein; light pink, hypothetical protein
Characterization of the pckA gene
The nucleotide sequence of ORF1 showed 91 % identity with the chvI gene sequence, however, the deduced protein product of the ORF showed 97 % homology with the transcriptional regulatory protein (ChvI) of A. tumefaciens C58. chvI codes for a response regulator of a two-component regulatory system chvGI (Charles and Nester 1993). ORF2 was the pckA gene of 1,611 bp that encoded PCK consisting of 536 amino acids as was also found in A.tumefaciens (Liu et al. 2005). The deduced protein product shared 96, 81, 80, 78 and 77 % identity with that of A. tumefaciens C58, R. etli CFN42, R. vitis S4, E. medicae WSM419 and E. meliloti 1,021, respectively. The pckA is classified as an acid-inducible gene under the control of chvGI, and is important in virulence of A.tumefaciens (Charles and Nester 1993). ORF3 was located downstream of the pckA gene and deduced protein product of the ORF shared 97 % homology with a conserved hypothetical protein of A. tumefaciens C58. All the open reading frames, except the ORF1, were transcribed in the same direction (Fig. 1a).
The genomic context of pckA gene in R. pusense, as compared to those in the related members of Rhizobiales showed that it was almost identical to that of R. radiobacter, although considerable homogeneity in composition with those of E. meliloti, Mesorhizobium loti and R. etli was also observed. All the four genomes were found to have gene orthologs for two component response regulator and phosphoenol pyruvate carboxykinase in their genome (Fig. 1b).
The pckA gene of 1,611 bp was amplified from R. pusense NRCPB100 by PCR using the primers designed from the flanking region of the ORF2 of NRC43. The amplicon was cloned into pGEM-TEasy vector, the recombinant plasmid kSD2 was isolated and the pckA in the clone was sequenced. The location of the Tn5 insertion was determined from the sequence of the transposon insertion junction in the pckA gene. It was between nucleotides 436 and 438.
A stretch of 1.851 kb DNA from R. pusense NRCPB100 containing pckA and its promoter sequence was also amplified by PCR using primers designed from the flanking regions of the ORF2 of NRC43. The amplicon was cloned into the broad host range vector pSUP104 to yield kSD3. The promoter sequence was included for expression of pckA gene. Complementation of the mutant R. pusense NRC43 with the plasmid kSD3 resulted in a phenotype similar to that of its parent in respect of growth on succinate, malate, fumarate or pyruvate. The mutant NRC43 did not grow on glucogenic substrates such as C4-dicarboxylic acids as the sole carbon source. This suggested that an alternative pathway for synthesis of PEP (Hansen and Juni 1974) did not exist in R. pusense NRCPB100 as was also found in A. tumefaciens C58 (Liu et al. 2005). The results confirmed that in NRC43 pckA gene was biologically non-functional and complementation with pckA restored its function and gluconeogenic activity.
When genomic DNA from R. pusense NRC43 was digested with EcoRI and hybridized with ~1.2 kb 32P-labeled neomycin gene of Tn5 as the probe a positive signal was obtained on a 8.5 kb DNA fragment of the strain NRC43 (Fig. 2a). To examine the location of Tn5, genomic DNA of NRCPB100 or NRC43 was digested with EcoRI and hybridized with the 1,611 bp pckA gene probe amplified by PCR from R. pusense NRCPB100 DNA. This resulted in the development of only one hybridization band signal on a EcoRI DNA fragment of about 8.5 kb of NRC43 or on a 2.737 kb EcoRI DNA fragment of NRCPB100 (Fig. 2b). The data demonstrated that there was only a single site of Tn5 insertion in the chromosomal DNA of NRC43. Since Tn5 does not have any EcoRI site on it, insertion of 5.8 kb Tn5 in the 2.737 kb EcoRI DNA fragment of NRCPB100 increased its length to about 8.5 kb as observed in strain NRC43. From the data it was concluded that the phenotype of NRC43 was the result of insertion of Tn5 in the 2.737 kb EcoRI DNA fragment which caused the inactivation of the pckA gene.
Southern hybridization analysis to localize the site of Tn5 insertion in the strain NRC43 of Rhizobium pusense by hybridization using 1.2 kb DNA fragment containing neomycin gene as probe. a with EcoRI digested genomic DNA of NRCPB100 (Lane 1), and of NRC43 (Lane 2); bpckA containing DNA sequence by hybridization of PCR amplified 1.611 kb fragment containing the pckA gene from the wild type NRCPB100 as a probe with EcoRI digested genomic DNA of strain NRC43 (Lane 1), and of strain NRCPB100 (Lane 2)
Mutation in the pckA gene exerts profound biological effects. PCK− mutant of R. leguminosarum bv. viciae nodulated and fix N2 as effectively as its parent (McKay et al. 1985), while PCK− mutants of E. meliloti showed reduced level of N2 fixation (Finan et al. 1991). The PCK enzyme, as such, was thought to be important in infection and nodulation. A PCK mutant of Rhizobium sp. strain NGR234 exhibited a host dependent symbiotic phenotype while N2 fixation by the mutant was also appreciably affected depending on host plants (Østerås et al. 1991). A pckA mutant of A. tumefaciens was attenuated in virulence (Liu et al. 2005) and pckA-deficient M. bovis BCG was also attenuated in virulence (Liu et al. 2003). The observations tempted us to analyze pckA gene of R. pusense NRCPB10 for its possible role in the strain’s non-infectivity. However, the pckA gene and the derivative enzyme of it were found to have high identity with those of other members of Rhizobiales which are infective. The enzyme of the strain was also metabolically active.
Phylogenetic analysis
Phylogenetic analysis based on pckA gene sequence of R. pusense placed it in a cluster with pckA genes of other members of Rhizobiales (Fig. 3). Amino acid sequence alignment of the deduced enzyme showed that all of the putative functional regions were conserved in the ATP dependent PCK enzymes (Fig. 4). The PCK specific domain which is conserved in all ATP dependent PCKs analyzed to date was also found to be conserved in R. pusense enzyme (residues 192–201). In addition, kinase-1a (residues 236–243), kinase-2 (residues 253–257), the covalent metal ion binding site (residues 265–272) and ribose binding site (residues 276–285) were essentially identical among all the PCKs. On the other hand, more variability was observed in the adenine binding site, although the key residues, arginine and threonine (R441 and T447 in R. pusense NRCPB10), were identical to those of the others.
Neighbor joining tree showing the phylogenetic relationship of Rhizobium pusense (NRCPB100) with other related species based on 1,611 nucleotide bp of pckA gene sequence. Sequences showing similarity of more than 70 % only were considered in the phylogenetic tree. Bootstrap values based on 1,000 re-samplings are shown at the branch points. Bar, 2 nucleotide substitutions per 100 nucleotide position
Conservation of the active site residues of ATP-dependent PCKs. Identical residues are indicated with asterisks below the sequence alignment. Rhizobium pusense (accession no: AAN86775); Bacillus subtilis (accession no: NP_390934); Pseudomonas aeruginosa (accession no: NP_253879); Sinorhizobium meliloti (accession no: NP_384151); Mesorhizobium loti (accession no: NP_105818); Escherichia coli (accession no: NP_417862) and Salmonella enterica (accession no: NP_458404). In the sequence of PCK of R. pusense the residue numbers are indicated immediately right to the residues
Conclusion
A pckA mutant NRC43 of the recently described non-nodulating and non-tumorigenic species R. pusense NRCPB10 failed to grow on succinate. The pckA gene of 1,611 bp encoding PCK was located between the transcriptional regulatory protein gene chvI and a conserved hypothetical protein gene. The derivative protein of the gene, a PCK enzyme, had high degree of homology with the known ATP-dependent PCKs and all the putative functional regions of the enzyme were conserved. Phylogeny of the pckA gene placed the strain in a cluster with other known members of Rhizobiales. The pckA gene was functional in R. pusense NRCPB10 and was induced on glucogenic substrates indicating that the regulation mechanism of the gene was also functional. Non-infectivity of R. pusense NRCPB10 was not, thus, due to a deficiency in pckA gene activity but for some other factor which was not clear in this study.
References
Altschul SF, Madde TL, Schaffer AA, Zhang J, Zheng Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Bluin J-M, Bortolis S, Nacfor M, Collinet M, Penot G, Laurent-puig P, Forest C (2010) Down regulation of the phosphoenolpyruvate carboxykinase gene in human colon and induction by omega-3 fatty acids. Biochimie 92:1772–1777
Charles TC, Nester EW (1993) A chromosomally encoded two-component sensory transduction system is required for virulence of Agrobacterium tumefaciens. J Bacteriol 175:6614–6625
Ciria R, Abreu-Goodger C, Morett E, Merino E (2004) GeConT: gene context analysis. Bioinformatics 14:2307–2308
Cooper RA, Kornberg HL (1967) The direct synthesis of phosphoenolpyruvate from pyruvate by Escherichia coli. Proc Royal Soc London B Biol Science 168:263–280
Felsenstein J (1985) Confidence limits on phylogenies: an approach using the bootstrap. Evolution 39:783–791
Figurski DH, Helinski DR (1979) Replication of an origin containing derivative of plasmid RK2 dependent on a plasmid function provided in trans. Proc Nat Acad Sci USA 76:1648–1652
Finan TM, Oresnik I, Bottacin A (1988) Mutants of Rhizobium meliloti defective in succinate metabolism. J Bacteriol 170:3396–3404
Finan TM, McWhinnie E, Driscoll B, Watson RJ (1991) Complex symbiotic phenotypes result from gluconeogenic mutations in Rhizobium meliloti. Mol Plant Microbe Interact 4:386–392
Gautam US, Jajoo A, Singh A, Chakrabartty PK, Das SK (2007) Characterization of an rpo mutant of Mesorhizobium ciceri. J Appl Microbiol 103:1798–1807
Goldie H (1984) Regulation of transcription of the Escherichia coli phosphoenolpyruvate carboxykinase locus: studies with pck-lacZ operon fusions. J Bacteriol 159:832–836
Hansen EJ, Juni E (1974) Two routes for synthesis of phosphoenolpyruvate from C4-dicarboxylic acids. Biochem Biophy Res Commun 59:1204–1210
Kimura M (1980) A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotides sequences. J Mol Evol 16:111–120
Liu K, Yu J, Russel DG (2003) pckA-deficient Mycobacterium bovis BCG shows attenuated virulence in mice and macrophages. Microbiol 149:1829–1835
Liu P, Wood D, Nester EW (2005) Phosphoenolpyruvate carboxykinase is an acid-induced, chromosomally encoded virulence factor in Agrobacterium tumefaciens. J Bacteriol 187:6039–6045
McKay IA, Glenn AR, Dilworth MJ (1985) Gluconeogenesis in Rhizobium leguminosarum MNF3841. J Gen Microbiol 131:2067–2073
McKay IA, Dilworth MJ, Glenn AR (1988) C4-Dicarboxylate metabolism in free-living and bacteroid forms of Rhizobium leguminosarum MNF3841. J Gen Microbiol 134:1433–1440
Meade HM, Long SR, Ruvkun GB, Brown SE, Ausubel FM (1982) Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobiummeliloti induced by transposon Tn5 mutagenesis. J Bacteriol 149:114–122
Merrero I, Rhee KY, Schnappinger D, Pathe K, Ehrt S (2010) Gluconeogenic carbon flow of tricarboxylic acid cycle intermediates is critical for Mycobacterium tuberculosis to establish and maintain infection. Proc Natl Acad Sci USA 107:9819–9824
Miller JH (1972) Experiments in molecular genetics. Cold Spring Harbor Laboratory, Cold Spring Harbor
Østerås M, Finan TM, Stanley J (1991) Site directed mutagenesis and DNA sequence of pckA of Rhizobium NGR234, encoding phosphoenolpyruvate carboxykinase: gluconeogenesis and host-dependent symbiotic phenotype. Mol Gen Genet 230:257–269
Østerås M, Driscoll BT, Finan TM (1995) Molecular and expression analysis of the Rhizobium meliloti phosphoenolpyruvate carboxykinase (pckA) gene. J Bacteriol 177:1452–1460
Østerås M, O’Brien SAP, Finan TM (1997) Genetic analysis of mutations affecting pckA regulation in Rhizobium (Sinorhizobium) meliloti. Genetics 147:1521–1531
Panday D, Schuman P, Das SK (2011) Rhizobium pusense sp. nov., isolated from rhizosphere of chickpea (Cicer arietinum L). Int J Syst Evol Microbiol 61:2632–2639
Priefer UB, Simon R, Puhler A (1985) Extension of the host range of Escherichia coli vectors by incorporation of RSF1010 replication and mobilization functions. J Bacteriol 163:324–330
Ronson CW, Lyttleton P, Robertson JG (1981) C(4)-dicarboxylate transport mutants of Rhizobium trifolii form ineffective nodules on Trifolium repens. Proc Nat Acad Sci USA 78:4284–4288
Sambrook J, Russell DW (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory, Cold Spring Harbor
Selvaraj G, Iyer VN (1983) Suicide plasmid vehicles for insertion mutagenesis in Rhizobium meliloti and related bacteria. J Bacteriol 156:1292–1300
Southern EM (1975) Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol 98:503–517
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 24:1596–1599
Utter MF, Kolenbrander HM (1972) Formation of oxaloacetate by CO2 fixation on phosphoenolpyruvate. In: Boyer PD (ed) The enzymes, vol VI. Academic Press, New York, pp 117–170
Vincent JM (1970) A manual for the practical study of the root nodule bacteria. I.B.P. Hand book No.15, Blackwell, Oxford
Voet D, Voet J, Pratt C (2008) Fundamentals of biochemistry. Wiley, New York
Wood WB (1996) Host specificity of DNA produced by Escherichia coli: bacterial mutations affecting the restriction and modification of DNA. J Mol Biol 16:118–133
Yang J, Kalhan SC, Hansen RW (2009) What is the metabolic role of phosphoenolpyruvate carboxykinase? J Biol Chem 284:27025–27029
Young JM, Kuykendall LD, Martinez-Romero E, Kerr A, Sawada H (2001) A revision of Rhizobium Frank 1889, with an emended description of the genus, and the inclusion of all species of Agrobacterium Conn 1942 and Allorhizobium undicola de Lajudie et al. 1998 as new combinations: Rhizobium radiobacter, R. rhizogenes, R. undicola and R. vitis. Int J Syst Evol Microbiol 51:98–103
Acknowledgment
This work was supported by a grant from the Department of Biotechnology, Ministry of Science and Technology, Government of India.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Das, S.K., Chakrabartty, P.K. Characterization of a (pckA) mutant of the non-nodulating bacterium Rhizobium pusense NRCPB10 induced by transposon Tn5 mutagenesis. 3 Biotech 2, 311–318 (2012). https://doi.org/10.1007/s13205-012-0057-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13205-012-0057-5