Abstract
A potential biosurfactant producing isolate was identified as Bacillus aryabhattai strain ZDY2. Biosurfactant production was enhanced by 2.51-fold through the development of an optimized process using response surface methodology. The optimized culture medium contained crude oil 4.0%, yeast extract 0.7% and NaNO3 3.0% that yielded 8.86 g/l of biosurfactant. Biosurfactant was characterized for stability up to 100 °C, at pH 5–10 and in the presence of NaCl concentration up to 8%. Biosurfactant demonstrated antimicrobial activity against Salmonella typhimurium, Escherichia coli, Micrococcus luteus, Staphylococcus aureus and Candida tropicalis. The morphological characterization was carried out by scanning electron microscopy with energy-dispersive X-ray analysis. The Fourier-transform infrared spectroscopy analysis reveals the lipopeptide nature of the biosurfactant produced by B. aryabhattai strain ZDY2. The biosurfactant finds application in healthcare and pharmaceutical industries.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The increasing trends on the application of biosurfactant in various industries have led to the worldwide interest on biosurfactant research. The microbial biosurfactants have been proved to be more environmental friendly over the chemical surfactants (Paraszkiewicz et al. 2018). Biosurfactants are comparatively nontoxic and biodegradable products with unique properties such as superior foaming ability, high selectivity and active under adverse environmental conditions of pH, temperatures and salinity (Varjani and Upasani 2017). The microbial biosurfactants are amphipathic in nature with hydrophobic and hydrophilic regions that permit them to partition at the interface between immiscible liquid phases with diverse polarities such as oil and water (Ohadi et al. 2017). Based on the origin and chemical nature, the biosurfactants are largely classified as lipopeptides, lipoproteins, glycolipids, phospholipids, fatty acids and polymeric molecules (Varjani and Upasani 2017). Furthermore, based on the molecular weight, biosurfactants are grouped as low molecular weight compounds (rhamnolipids, sophorolipids, lipopeptides and trehalolipids) and high molecular weight compounds (lipoprotein and polymeric molecules) (Elshikh et al. 2016). However, glycolipids and lipoproteins have gained preference in industries (Geys et al. 2014) and glycolipids nature of biosurfactant has to be reported by researchers (Astuti et al. 2019; Dwivedia et al. 2019). These specific groups of biosurfactants display a wide spectrum of antimicrobial properties and hence are exploited for biomedical activities, cosmetic, food and pharmaceutical applications (Elshikh et al. 2016; Ndlovu et al. 2017). Lipopeptides constitute of amino acid chain forming the peptide moiety (linear or cyclic) and varying length of fatty acid. Commonly found antimicrobial lipopeptides include surfactin, subtilisin, lichenysin, viscosin, serrawettin, fengycin, arthrofactin, etc. (Shekhar et al. 2014). Biosurfactants have widespread applications in enhanced oil recovery, biodegradation industries, in sustainable agriculture as fungicides, insecticides and herbicides, moistening, dispersing, emulsifiers, foaming agents and healthcare (Sachdev and Cameotra 2013; Barakat et al. 2017; Varjani and Upasani 2017). Microbial genera reported for biosurfactant production include Acinetobacter, Arthrobacter, Pseudomonas, Halomonas, Bacillus, Rhodococcus, Enterobacter, yeast species, etc. Among them, Bacillus species are commonly found to produce lipopeptides (Shekhar et al. 2014). Large-scale production of biosurfactants for different industrial applications is always challenging (Makkar et al. 2011). Optimization of production parameters greatly influences the biosurfactant productivity. Several optimization strategies such as one factor at a time and statistical designs like factorial designs and response surface methodology have been adopted for biosurfactant optimization (Almansoory et al. 2017; Ohadi et al. 2017; Souza et al. 2018).
However, fewer reports are available on biosurfactant production and optimization from Bacillus aryabhattai strain. The current study focused on the statistical optimization and characterization of antimicrobial lipopeptide biosurfactant molecule produced by isolated B. aryabhattai strain ZDY2. The stability of biosurfactant at different environmental conditions was assessed.
Materials and methods
Chemicals and strains
All the chemicals and standards used in the present study were obtained from Merck and Co., Inc. (USA) and Sigma-Aldrich Pvt. Ltd. (USA). Escherichia coli MTCC 443, Salmonella typhimurium MTCC 98, Pseudomonas aeruginosa MTCC 2297, Micrococcus luteus NCIM 2871, S. aureus MTCC 737, Bacillus cereus NCIM 2217 and isolated yeast Candida tropicalis cultures were used for antimicrobial studies. The cultures were procured from Microbial Type Culture Collection and Gene Bank (MTCC), Chandigarh, and National Collection of Industrial Microorganisms (NCIM), Pune.
Isolation and screening of biosurfactant producing bacteria
Crude oil contaminated sites were explored for the isolation process. Soil samples from petroleum refineries with oil spillages were collected from regions of Hubballi, Karnataka, India. The soil samples were collected from 10–15 cm depth in sterile polythene bags and stored in laboratory at room temperature (25 ± 2 °C) for isolation. Isolation was carried out by enrichment technique. The soil sample was enriched in minimal salt medium (MSM) containing (g/l) NaNO3 7.5; KCl 1.1; NaCl 1.1; FeSO4·7H2O 0.00028; K2HPO4 3.4; MgSO4. 7H2O 0.5; yeast extract 0.5; glucose 10.0 and trace element solution (g/l) ZnSO4·7H2O 0.29; CaCl2·4H2O 0.24; CuSO4·5H2O 0.25; and MnSO4·H2O 0.17 supplemented with 1% crude oil at pH 7.0 (Joy et al. 2017). The flasks were incubated for 5–6 days at 30 ± 2 °C in orbital shaker (60 rpm). The enriched culture was plated on MSM agar media supplemented with 1% crude oil by serial dilution. Plates were incubated at 30 ± 2 °C for 48 h. Morphologically distinct isolated colonies were purified by repeated streaking. The isolated pure bacteria capable of degrading crude oil based on their growth were maintained on MSM agar slant and stored at 4 °C until further use. The pure cultures were primarily screened for biosurfactant producers on modified cetyltriammonium bromide (CTAB) methylene blue agar media with the following composition (g/l) CTAB 5.0; methylene blue 0.02; beef extract 3.0; peptone 10.0; yeast extract 0.1; NaCl 2.0; and agar 20.0 (Liu et al. 2017). The culture showing dark blue halo was considered as positive biosurfactant producers. The isolates were also screened for its biosurfactant potential on blood agar plate containing (g/l) trypticase 10.0; beef extract 3.0; NaCl 5.0; and agar 20.0 supplemented with 5% sheep blood. The zone of clearance around the colony indicates the hemolysis. The positive isolate (ZDY2) showing highest zone was further assessed for biosurfactant production (Ohadi et al. 2017).
Growth conditions for production of biosurfactant
Pre-inoculum of the pure positive isolate ZDY2 was prepared in 50 ml of the previously described MSM liquid media supplemented with 1% crude oil and incubated at 30 ± 2 °C and pH 7.0 for 24 h with agitation (150 rpm). 10% of this inoculum (OD600 0.85) was transferred to the 250-ml Erlenmeyer flask contained 100 ml of MSM production media supplemented with 1% crude oil. Flask was incubated under similar conditions as described above for 5 days. The culture broth was centrifuged at 8000 rpm for 15 min at 4 °C to obtain a cell-free supernatant which was used for the analysis of biosurfactant activities.
Assessment of biosurfactant activity
The cell-free supernatant of the positive isolate (ZDY2) was assessed for biosurfactant activities by following various assays. The reduction in the surface tension between hydrophilic and hydrophobic surface by the biosurfactant was assessed. Drop-collapse test was performed by placing 25 μl of the cell-free supernatant in the form of drop over a hydrophobic surface of parafilm strip. The occurrence of biosurfactant in the supernatant reduces the surface tension between the drop and hydrophobic surface causing the drop to collapse. Supernatant of un-inoculated media was used as a negative control (Joy et al. 2017). Oil spreading assay was carried out as follows. Approximately 40 ml of distilled water was taken in a Petri plate, and 25 μl of mineral oil was overlaid over the water surface. Ten microliters of the cell-free supernatant was added to the surface of mineral oil, and the displacement of mineral oil by the biosurfactant was measured (Ohadi et al. 2017). Emulsification index (E24) of the biosurfactant was determined by adding 2.0 ml diesel oil (hydrocarbon) to 2.0 ml of cell-free supernatant. After vigorous mixing at high speed, it was allowed to stand for 24 h. Emulsification index (E24) percentage was calculated by measuring the height of the emulsified layer (mm) divided by the total height of the solution (mm) (Derguine-Mecheri et al. 2018). The emulsification activity (EU/ml) was determined by measuring the absorbance (400 nm) of the aqueous layer obtained from the emulsion of diesel oil (0.5 ml) and cell-free supernatant (3.0 ml) according to method described by Khopade et al. (2012). To determine the cell surface hydrophobicity of the strain, the strain ZDY1 was grown for 4d in MSM liquid media supplemented with 1% crude oil. The cells were harvested, and the OD600 was adjusted to 0.5. Crude oil was added to the cell suspension, and the changes in OD600 were measured. The cell surface hydrophobicity of the strain was calculated according to Kumari et al. (2012).
Molecular identification of potent biosurfactant producing bacteria by 16S rRNA gene sequencing
Molecular identification of the isolated biosurfactant producing bacterial strain was carried out by 16S rRNA gene sequence analysis. Genomic DNA was isolated from the pure culture and used for 16S rRNA gene amplification by PCR. Amplification of the 16S rRNA gene was carried out using the universal primers with a defined PCR reaction as described by Mulla et al. (2016, 2018). The amplified PCR product was gel purified and sequenced. The obtained nucleotide sequence was analyzed and aligned with those of the closely related gene sequences available in the GenBank database using the BLAST-n and CLUSTAL-W program. The phylogenetic tree was build based on the maximum likelihood method with 1000 bootstrap replicates using MEGA6 software (Tamura et al. 2013).
Extraction of biosurfactant
Acid precipitation and solvent extraction method was adopted for the extraction of biosurfactant. The culture broth (5d incubated) was centrifuged at 8000 rpm for 15 min at 4 °C. The cell-free supernatant was adjusted to pH 2.0 with 6 N HCl and subjected to precipitation overnight under cold condition (4 °C). This was followed by liquid–liquid extraction by chloroform/methanol (65:15). The biosurfactant was concentrated using rotary evaporator (45 °C). The concentrated biosurfactant was freeze-dried by lyophilizer, and yield (dry weight) was expressed in g/l (Pruthi and Cameotra 1997).
Biosurfactant production kinetics
The biosurfactant production kinetics was studied by estimating the dry cell biomass and biosurfactant production in the fermentation (batch) broth over the time period. Pre-inoculum was prepared and inoculated into MSM (1% crude oil) as described above. The flasks were incubated for 6 days, and aliquots were withdrawn aseptically at regular intervals of 24 h from the fermentation broth and centrifuged (8000 rpm, 15 min, 4 °C) to obtain a cell pellet. The pellet was washed twice with distilled water and dried to obtain constant weight. The biosurfactant was extracted from the supernatant as described above. The dry cell biomass and biosurfactant yield were expressed in g/l (Khopade et al. 2012; Santos et al. 2017).
Effect of culture conditions on biosurfactant production
The effects of different culture conditions on biosurfactant production (g/l) were evaluated. To study the effect of the substrate, crude oil, diesel, olive oil and glycerol (1% v/v) were supplemented in the MSM. Temperature (25, 30, 35 and 40 °C) and pH (5.0, 6.0, 7.0, 8.0 and 9.0) effect was analyzed. The biosurfactant production was also evaluated under agitation (200 and 300 rpm) and static conditions. Biosurfactant was extracted, and emulsification index (E24) percentage was calculated as described above.
Statistical design for optimization of biosurfactant production
Selection of critical parameters by PBD
The critical parameters influencing the biosurfactant production were screening by Plackett–Burman design (PBD) after analyzing the effects of different culture conditions during preliminary study. PBD is a two-level fractional design employed to investigate ‘n’ factors in n + 1 experiments assuming no interactions exist between the factors (Plackett and Burman 1946). Eight factors were selected for screening, namely NaNO3 (X1), KCl (X2), NaCl (X3), KH2PO4 (X4), MgSO4·7H2O (X5), yeast extract (X6), glucose (X7) and crude oil (X8). Minitab 17 statistical software was used to create the design matrix and result analysis. Table 1 represents 12 experiment trails generated at two levels high (+ 1) and low (− 1) with the actual and coded values of factors and their respective mean responses as biosurfactant yield (g/l). Biosurfactant production was executed at pH 7.0 and temperature 35 °C for 4 days with agitation at 200 rpm. Experiments were conducted in duplicates, and interpretation of the responses followed the first-order polynomial model equation (Bagewadi et al. 2018); Y is the response (biosurfactant yield g/l). Significant effect of critical parameter was depicted from the probability ‘p’ value of factors.
Optimization of biosurfactant yield by RSM design
To further improve the biosurfactant yield, central composite design (CCD) of response surface methodology (RSM) was adopted. The significant factors analyzed by PBD were chosen for optimization. Minitab 17 statistical software was used to create the design matrix and for statistical analysis. CCD was implemented to scrutinize the most optimum level, impact and interactions of three factors such as crude oil (A), yeast extract (B) and NaNO3 (C) on biosurfactant production. The factors were assessed at five coded levels (− α, − 1, 0, + 1 and + α), and their actual and coded values are shown in Table 2. The design matrix comprising of 20 experiments in duplicates is shown in Table 2. The other media constituents were kept constant. The correlation between the coded values of the input variables and the actual values was defined in equation described elsewhere (Bagewadi et al. 2018). The biosurfactant yield (g/l) corresponds to the response (Y) as shown in Table 2. A second-order polynomial quadratic (Eq. (2)) was fitted to response as follows:
where Y represents the response (biosurfactant yield g/l), A, B and C are the independent factors, β1, β2 and β3 are the linear regression coefficients, β11, β22 and β33 are the quadratic coefficients, and β12, β13 and β23 are the cross-product coefficients of the model (Bagewadi et al. 2016).
The response surfaces plots of the factors were obtained using Minitab 17 statistical software to study the interactions among the factors. Analysis of variance (ANOVA) was applied to the response (biosurfactant yield g/l) obtained from the optimization design. ANOVA was used to check the interaction effects and significance of the factors influencing the process of biosurfactant production. The polynomial model equation was assessed with the obtained values coefficient of determination R2 and adjusted R2. The probability ‘p’ and Fisher’s ‘F’ function relate to the model significance. Validation of the obtained model was verified experimentally (triplicates) under optimized condition. For the optimization studies, the biosurfactant (g/l) was extracted as described above.
Stability of biosurfactant
Stability of biosurfactant was assessed under different conditions and expressed as emulsification index (E24) percentage as described above. The cell-free supernatant was obtained by centrifuging at 8000 rpm for 15 min (4 °C). Thermal stability of biosurfactant was estimated by maintaining the cell-free supernatant at constant temperature between 30 and 100 °C for 30 min and then cooled to room temperature. The pH effect was evaluated in the range of pH 2–10 (1 M HCl or 1 M NaOH), and salinity was assessed using NaCl (2–10% w/v). All the assays were carried out in triplicates (Mouafi et al. 2016; Chebbi et al. 2017).
Assessment of antimicrobial properties of biosurfactant
In vitro antimicrobial activity of extracted biosurfactant (5.0 mg/ml) was evaluated against Gram-negative bacteria such as E. coli, S. typhimurium and Pseudomonas aeruginosa and Gram-positive bacteria such as M. luteus, S. aureus and B. cereus. Biosurfactant was also tested against yeast C. tropicalis by agar well diffusion method described previously by Bagewadi et al. (2019). The antimicrobial activity was assessed on the basis of zone of inhibition (mm). Gentamicin (1 mg/ml) and DMSO served as positive and negative control, respectively.
Cytotoxicity assessment
The cytotoxicity of extracted biosurfactant was evaluated on the human lung carcinoma (A549) cell line by [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide] (MTT) assay according to the method described by Bhat et al. (2018). The absorbance measurements were recorded at 570 nm using a microplate reader. The biosurfactant was tested at a concentration of 12.5, 25, 50 and 100 mg/ml. The dimethyl sulfoxide (DMSO) was used as solvent and was treated as a negative control in the assay, and triton was used as positive control. The growth inhibition in terms of cell viability percentage was calculated. All the experiments were conducted in triplicate.
Analytical characterization
FTIR analysis
FTIR (PerkinElmer, FTIR1760) analysis was carried out to explore the presence of chemical functional groups in biosurfactant according to previous report (Bagewadi et al. 2017). The extracted biosurfactant was freeze-dried by lyophilizer and used for preparation of KBr (potassium bromide) for analysis in scanning range of 500–4000 cm−1.
Scanning electron microscopy (SEM) with energy-dispersive X-ray (EDX) analysis
The morphological nature of produced biosurfactant was studied by SEM–EDS (VEGA/TESCAN, USA), and elemental analysis was carried out by energy-dispersive X-ray microanalysis system of SEM according to previously published method (Bagewadi et al. 2016).
Results and discussion
Isolation, screening and molecular analysis by 16S rDNA gene sequencing of biosurfactant producer
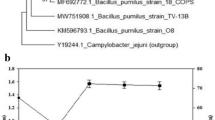
Prospective biosurfactant producers were isolated from petroleum refineries inhabiting crude oil spillages. Isolation was effectively carried out by enrichment technique using MSM supplemented with 1% crude oil. Among the isolated bacterial isolates, ZDY2 strain demonstrated to be a potential biosurfactant producer showing colony with dark blue halos. ZDY2 strain was also subjected for preliminary screening on blood agar plate, which showed hemolytic activity with a highest zone of 12 mm (data not shown). The positive strain ZDY2 was further confirmed for biosurfactant production by assessing biosurfactant activities. To evaluate the biosurfactant efficiency of the isolates, the strategy of combination of different screening assays was adopted. The strain showed positive for drop-collapse test with the formation of completely flat drop due to the reduction in the surface tension. Further the strain showed the displacement of mineral oil with a diameter of 10 mm during the oil spreading assay. Furthermore, the emulsification index (E24), emulsification activity and cell surface hydrophobicity were found to be 68 ± 0.5%, 124 EU/ml and 60 ± 1.3%, respectively. The results supported the potentiality of the strain for biosurfactant production. Isolated strain ZDY2 was molecularly identified by 16S rRNA gene sequencing and phylogenetic analysis. The isolated genomic DNA showed a single distinct high molecular weight DNA band which was amplified to obtain the PCR amplicon band. The PCR amplicon was purified and sequenced. Nucleotide BLAST (BLAST-n) was performed for the obtained consensus sequence to compare maximum likelihood with the available sequences at National Center for Biotechnology Information (NCBI) GenBank database. The sequences were aligned using CLUSTAL-W. The phylogenetic analysis was carried out using MEGA6 software as shown in Fig. 1. Based on the phylogenetic analysis, the strain was identified as B. aryabhattai species and designated as B. aryabhattai strain ZDY2. The identified strain sequence has been deposited to NCBI databank with the accession number MK508863. Several workers have exploited various environmental ecosystems such as soil, water and plants for biosurfactant producing bacteria (Sachdev and Cameotra 2013). Other researchers have reported the isolation of biosurfactant producing bacteria such as Pseudomonas taiwanensis L1011 (crude oil sample) (Liu et al. 2017), Acinetobacter junii B6 (soil sample) (Ohadi et al. 2017), Bacillus amyloliquefaciens SH20 and Bacillus thuringiensis SH24 (Red Sea, Egypt) (Barakat et al. 2017). However, not many reports are available on biosurfactant producing B. aryabhattai strain. Biosurfactant produced by B. aryabhattai strain ZDY2 can be employed for cleaning of oil spillages.
Kinetics of biosurfactant production
Kinetic models are important techniques for the process development and industrial application (Heryani and Putra 2017). Production profile predicts the maximum productivity period. The biosurfactant production kinetics revealed that B. aryabhattai strain ZDY2 grew well in MSM containing crude oil. The strain was able to utilize glucose as the source of carbon and crude oil as inducing substrate for the production of biosurfactant. Biosurfactant production initiated after 24 h. Maximum biosurfactant production of 3.52 g/l occurred on fourth day of fermentation with cell biomass of 1.3 g/l (Fig. 2). After fifth day, the biosurfactant production and cell growth declined. The biosurfactant production by the strain was growth dependent indicating that the produced biosurfactant is a primary metabolite. The decrease in the cell biomass after high product concentration is a common observation during a fermentation process. Similar growth-associated biosurfactant production has been also reported from Virgibacillus salarius (KSA-T) (Elazzazy et al. 2015). Ismail et al. (2015) studied biosurfactant production kinetics in P. aeruginosa AK6U strain using MSM with glucose as carbon and 4,6-dimethyldibenzothiophene as sulfur source. However, Heryani and Putra (2017) revealed the kinetics in Bacillus sp. by adopting the Gompertz equation to predict the cell mass and biosurfactant production as a goodness of fit. The biosurfactants may function for the survival of the microorganisms by supporting transport of nutrients. The produced biosurfactant also enhances the utilization of provided hydrocarbon, in this case the crude oil, and assists in cell growth.
Effect of culture conditions on biosurfactant production
The production of biosurfactant is governed by several factors such as strains, production media components (carbon and nitrogen), salt concentrations, pH, temperature, agitation speed and aeration (Banat et al. 2014; Almansoory et al. 2017). Supplementation of carbon source in the medium is an essential component for enhancement of biosurfactant productivity. All the four carbon sources supported the production of biosurfactant. Crude oil was found to be a better carbon source to support the biosurfactant production of 3.52 g/l with emulsification index (E24) of 74% as shown in Fig. 3a. Olive oil (3.3 g/l) and diesel (3.2 g/l) also induced efficient production. However, considering the economics of production, crude oil and diesel are better sources. Glycerol did not efficiently support the growth of the strain and help result in low productivity (2.4 g/l). As shown in Fig. 3b, B. aryabhattai strain ZDY2 produced 4.0 g/l and 4.2 g/l of biosurfactant with E24 of 77% and 66% at 35 °C and 40 °C, respectively. Based on the emulsification activity, 35 °C was considered optimum. Increase in temperature conditions elevated the biosurfactant productivity; however, at temperatures beyond 45 °C, the growth of the strain was hampered yielding low biosurfactant (could not be quantified). pH also influences the biosurfactant production as shown in Fig. 3c. Biosurfactant production was maximum (4.0 g/l) between pH 7.0 and 8.0 with E24 of 78%. Alkaline condition beyond pH 9.0 decreased the productivity drastically. Under acidic condition, there was a profound reduction in the productivity indicating the sensitivity of microbial growth at lower pH. Similarly, Stenotrophomonas maltophilia NBS-11 also showed maximum biosurfactant production at pH 7.0 (Hemlata et al. 2015). Figure 3d represents the influence of different agitation and static conditions on production. The highest production of 4.3 g/l with E24 of 80% was achieved at 200 rpm. At higher rpm of 300, the productivity reduced by 50%. However, static condition did not support any significant biosurfactant production (0.8 g/l). The significance of oxygenation for biosurfactant production is well known, and previous report suggests that increase in the agitation had a negative effect on the biosurfactant production by Candida lipolytica (Santos et al. 2014). In comparison with our results, Almansoory et al. (2017) reported glycerol as best carbon source with biosurfactant production of 1.05 g/l, temperature of 30 °C and agitation speed of 200 rpm from Serratia marcescens.
Statistical optimization of biosurfactant production from B. aryabhattai strain ZDY2
Screening of significant parameters affecting biosurfactant production
Nutritional requirement for microbial growth and product production varies among the species. Hence, design of specific requirements is important to improve the productivity. Improvement of a process necessitates optimization of conditions involving several parameters such as media composition in order to yield high biosurfactant production. Plackett–Burman design is regularly used to screen out significant factors and estimate their main effects. The PBD results reveal the effect of significant factors on biosurfactant production by B. aryabhattai strain ZDY2. The experimentally obtained and predicted responses as biosurfactant production (g/l) are shown in Table 1. The sixth experimental run shows the maximum biosurfactant production of 5.88 g/l. The biosurfactant production ranges between 1.35 and 5.88 g/l among the 12 trails. The differences in the response values could be attributed to the influence of different combination levels of process parameters. Regression analysis and ANOVA with a probability ‘p’ < 0.05 determine the significance of the model terms. The large ‘F’ value and low ‘p’ are associated with greater significance of the relevant term. The model ‘F’ value (35.41) was significant. NaNO3 (X1), yeast extract (X6) and crude oil (X8) demonstrated a significant effect (‘p’ < 0.05) on biosurfactant production with the assessed concentration levels. Crude oil (p = 0.002; F = 116.68; Effect = 1.4850) had a greater positive influence on the biosurfactant production with larger ‘F’ and lower ‘p’ value. The significant effect of yeast extract (X6) and NaNO3 (X1) was also positive (effect of X6 = 1.40 and X1 = 1.01) as shown in Table 3. The coefficient of determination R2, adjusted R2 and R2 pred was 98.95%, 96.16% and 83.24%, respectively, that reflects a strong correlation between the observed and predicted responses. The first-order regression model was integrated to the obtained experimental data thereby illustrating the relationship between the independent variables and the response with the following equation.
where Y is the predicted response and X1, X2, X3, X4, X5, X6, X7 and X8 are the coded values of NaNO3, KCl, NaCl, KH2PO4, MgSO4.7H2, yeast extract, glucose and crude oil, respectively.
The PBD analysis reveals that crude oil has an enormous influence on biosurfactant production and used as a carbon source by the organism for production. The biosurfactant production is also influenced by the organic and inorganic nitrogen sources present in the media, and these are essential components for the growth of the bacteria. However, glucose and other minerals present in the media did not have a significant effect on the biosurfactant production. The lower concentration conditions of glucose in order to meet the initial growth requirements of bacteria and insignificant effects of minerals are well documented in the report by Nawawi et al. (2010). The significant components screened in PBD can be further optimized to study their influence on biosurfactant productivity. However, the previous studies conducted by Deepika et al. (2016) on biosurfactant production by P. aeruginosa strain KVD-HR42 using PBD also revealed that Karanja oil and sodium nitrate were found to have a significant effect. In agreement to our results, their results also suggest the insignificant effect of minerals. The results of Kumar et al. (2015) also showed the positive effects of sawdust, groundnut oil and glycerol for biosurfactant production by P. aeruginosa 2297. Other researchers found several factors such as dextrose, zinc chloride, magnesium sulfate and pH to exert a positive impact on production by S. maltophilia NBS-11 (Hemlata et al. 2015). Though the mutual interactive effect of the parameters is not suggested by PBD, it is basically adopted to locate the main effect of significant factors. Hence, this enables for further optimization studies.
Optimization of biosurfactant production by RSM design
RSM is a statistical technique used to analyze the correlation among the variables employed in the experimentation. RSM based on a CCD was applied to optimize the significant media components (crude oil, yeast extract and NaNO3) obtained from PBD in order to improvise the biosurfactant production by B. aryabhattai strain ZDY2. The investigational and predicted responses of 20 experimental trails are shown in Table 2. The investigational average response of 7.42 g/l was found to be highest with responses ranging between 1.1 and 7.4 g/l due to the variations in the concentration of the three selected factors. The highest response was obtained for the factors retained at ‘0’ coded level (crude oil 4%, yeast extract 0.5% and NaNO3 2.5%). ANOVA was adopted as it scrutinizes the variability produced by a factor. The model adequacy was tested from computed ‘F’ value of 24.74 and ‘p’ < 0.05 (Table 4). Regression analysis indicated crude oil (A) and NaNO3 (C) to be significant (‘p’ < 0.05) for biosurfactant production, whereas yeast extract (B) was found to be insignificant. The linear positive coefficient values of crude oil and NaNO3 indicate that these factors increase biosurfactant production. The results suggest that the occurrence of a hydrophobic substrate (crude oil) is essential for biosurfactant production. The interactions between the factors such as AB (crude oil–yeast extract; p = 0.028) and AC (crude oil–NaNO3; p = 0.004) were significant and among BC (yeast extract- NaNO3) were insignificant (Table 4). The insignificant interactions were expelled from the quadratic polynomial equation of the model. The correlation coefficient R2 is associated with model’s accuracy and was found to be 95.70%, and adjusted R2 was 91.83%. The obtained R2 implies greater correlation between the investigational and predicted responses. High consistency is indicated by smaller coefficient of variation. An insignificant lack of fit indicates the validation of quadratic model for the existing study. A second-order polynomial equation was used to ascertain the effect of factors on biosurfactant production by multiple regression analysis. The investigational data of combined effects of factors were fitted in Eq. (3) that explains the response (biosurfactant production g/l) based on the concentration of significant factors.
where Y is the response (biosurfactant production g/l) and A, B and C symbolizes the coded values of the independent factors.
To ascertain the optimum level of each independent factor for maximum productivity, 3D response surface plots were generated by plotting the response function (biosurfactant production g/l) of two factors while maintaining the other at central point. The significant interactions among the factors AB (crude oil–yeast extract) and AC (crude oil–NaNO3) were evidently obtained from the response surface plots. The center point concentrations of crude oil (4.0%) and 0.7% concentration of yeast extract demonstrate an increasing tendency in the biosurfactant production as shown in Fig. 4a. The crude oil of 4.0% is optimum for biosurfactant production, and beyond this concentration, there is a decline in the production probably due to hindrance of the bacterial growth due to increased viscosity in the medium with increase in crude oil concentration. The higher concentration of crude oil also affects the agitation of the medium and aeration to the organism. The yeast extract concentration beyond 0.7% depicts a decline in the production possibly due to production of biosurfactant under nitrogen-limiting conditions. A similar effect of yeast extract with optimum concentration of 4.6 g/l for biosurfactant production by Wickerhamomyces anomalus CCMA 0358 was reported (Souza et al. 2018). Analysis of a response plot (Fig. 4b) predicted high biosurfactant production with crude oil concentration between 3.0 and 4.0% as designated from the curvature of the response plot, and a decline in the response is observed below and above this concentration range. A linear increase in biosurfactant production was evidenced with increase in NaNO3 concentration up to 3.0% beyond which production remained constant. A balance of carbon/nitrogen ratio is important for optimum production of biosurfactant. NaNO3 being a source of nitrogen not only contributes for bacterial growth but is also involved as a structural component of biosurfactant specifically the lipopeptide biosurfactants (Nawawi et al. 2010). Based on the analysis, the subsequent optimal conditions for maximum biosurfactant production were designed, with crude oil 4.0%, yeast extract 0.7% and NaNO3 3.0%. Predicted optimization conditions necessitate verification, and hence, the optimal conditions were validated by experimentations carried out in triplicates. Biosurfactant production under optimized conditions was found to be 8.86 g/l in comparison with the un-optimized conditions (3.52 g/l). A 2.51-fold increase in the biosurfactant production was accomplished after optimization process. The obtained result suggests high accuracy of the developed model and model validation under the prescribed process conditions. Further, enhanced response can be achieved through improvement in optimization conditions by incorporating other critical physical process parameters. In this present research, we report a high biosurfactant production through optimization by B. aryabhattai strain ZDY2. Nevertheless, not much data are reported on optimization studies on biosurfactant production by B. aryabhattai strain. Employment of statistical optimization techniques helps to integrate and design processes to achieve high productivity. A number of workers have reported successful statistical optimizations of biosurfactant production from various bacteria such as optimization of biosurfactant production from Bacillus brevis using RSM (Mouafi et al. 2016), from Serratia rubidaea SNAU02 using mahua oil cake under solid-state fermentation (Nalini and Parthasarathi 2018), from C. lipolytica UCP0988 (Santos et al. 2014) and from Pseudozyma tsukubaensis using cassava wastewater (Fai et al. 2015). In P. aeruginosa strain KVD-HR42, biosurfactant production was optimized by Box–Behnken design of RSM using Karanja oil substrate which yielded 5.9 g/l of biosurfactant (Deepika et al. 2016).
Evaluation of biosurfactant stability and antimicrobial properties
Assessment of biosurfactant stability becomes critical for their application in various fields. The stability of biosurfactant from B. aryabhattai strain ZDY2 was evaluated at different temperature, pH and NaCl concentration conditions and expressed as emulsification index (E24) percentage. The biosurfactant showed thermal stability up to 100 °C with no significant change in the E24%. Greater than 90% of E24 was evidenced between 30 and 90 °C (Table 5). Results indicate wide thermal stability of the biosurfactant. Furthermore, the pH effect indicates the higher E24% at pH 7–8. There was no remarkable loss in the E24% between pH 5 and 10. However, at lower pH 2–3, there was a significant decrease observed in the E24% (Table 5). There is a tendency of the anionic biosurfactant to precipitate at lower pH values which is evidenced with a decreased E24%. The results reveal that the biosurfactant from cell-free supernatant is stable and retains its surface-active ability at pH range of 5–10. The ability of the biosurfactant to maintain surface-active properties at various ionic strengths was evaluated. The biosurfactant was stable in the presence of NaCl concentration up to 8% with E24 greater than 80%. Stability was compromised at higher NaCl concentration of 10% with decrease in E24 (75%) (Table 5). Thus, the stability assessment of the biosurfactant indicates its potential applications under varied environmental conditions. The stability profile of the biosurfactant is in agreement with previous reports. The variation in the stability and E24% of the biosurfactant under the influence of the temperature and pH was reported from B. brevis that showed thermal (30–80 °C) and pH (4–9) stability (Mouafi et al. 2016). In agreement of our results, decrease in the E24% at lower pH was also evidenced by Chebbi et al. (2017). Morais et al. (2017) also reported a decrease in the stability of biosurfactant produced by Lactobacillus jensenii P6A and Lactobacillus gasseri P65 with increased NaCl concentration. It is reported as a common tendency to lose surface-active properties at high salt concentration.
The extracted biosurfactant from B. aryabhattai strain ZDY2 was evaluated for in vitro antimicrobial potential against a range of microbial pathogens by agar well diffusion assay. The zone of inhibition (mm) for the tested pathogens is shown in Table 5. The biosurfactant demonstrated varying inhibitory action against both Gram-negative and Gram-positive microbial strains. The biosurfactant showed active inhibitory effect (> 10 mm) against all the tested pathogens. The results show that S. typhimurium (28 ± 0.5 mm) was most susceptible and P. aeruginosa (10 ± 0.9 mm) was least susceptible among the tested Gram-negative bacteria. An average zone of inhibition was observed for E. coli (15 ± 1.2 mm). The biosurfactant also revealed a significant inhibitory effect against M. luteus (22 ± 0.4 mm) and S. aureus (18 ± 0.8 mm) among the tested Gram-positive bacteria. Biosurfactant showed a pronounced effect against yeast C. tropicalis (25 ± 0.7 mm) (Table 5). The effects of positive (Gentamicin) and negative (DMSO) control were confirmed. The mechanism of action of biosurfactant, however, depends on the type of biosurfactant produced by the strain. Nevertheless, some hypothesis on the mechanism of antimicrobial action of biosurfactant was documented. The biosurfactant builds up at the microbial cell surface at higher threshold and penetrates into the cell by rupturing the cell membrane. The structural and functional alterations induced by biosurfactant cause cell death. Occurrence of various isoforms of glycolipids and lipopeptides signifies their structural heterogeneity (Ndlovu et al. 2017). A rhamnolipid type of biosurfactant having hydrophobic and hydrophilic groups can lead to substantial changes in the cell by inserting the fatty acid moiety in the cell membrane, and so also the acyl tails of the rhamnolipid bring about disintegration between the cytoskeleton elements and the plasma membrane. The surfactin and rhamnolipid extracts derived from B. amyloliquefaciens and P. aeruginosa have demonstrated pronounced antimicrobial activity against a broad spectrum of microorganisms, including S. aureus, E. coli and Candida albicans (Ndlovu et al. 2017). Another study reported antimicrobial activities against oral streptococci by biosurfactants produced by Lactobacillus spp. (Ciandrini et al. 2016). Morais et al. (2017) reported antimicrobial activity of biosurfactant from L. jensenii P6A and L. gasseri P65 against clinical isolates of E. coli, C. albicans, Staphylococcus saprophyticus, Enterobacter aerogenes and Klebsiella pneumonia. The studied antimicrobial potential of the biosurfactant can be employed against microbial strains which are resistant against chemical antibiotics.
Cytotoxicity assessment
The biosurfactant was assessed for cell-mediated cytotoxicity on the human lung carcinoma (A549) cell line by MTT assay. Figure 5 represents the cell viability percentage of cells treated with biosurfactant at varying concentration. The tested concentration of 12.5, 25, 50 and 100 mg/ml of biosurfactant did not show any significant growth inhibition. The cell viability percentage at 12.5 mg/ml and 100 mg/ml was 95.69% and 72%, respectively, indicating low toxicity of biosurfactant. However, increases in concentration of biosurfactant tend to reduce the cell viability %. The cell viability % of biosurfactant was higher compared to the triton (positive control). Hence, the effective concentration range of biosurfactant revealed lower toxicity suggesting the safety of extracted biosurfactant from B. aryabhattai strain ZDY2. Other reports also suggest low toxicity of the biosurfactant obtained from different sources. Sambanthamoorthy et al. (2014) reported the low toxicity of biosurfactant from Lactobacilli on human A549 lung epithelial cells at concentration of 25–100 mg/ml. Biosurfactant produced by Enterococcus faecium showed 90% cell viability at 6.25 mg/ml when evaluated for cytotoxicity on mouse fibroblast (ATCC L929) cell line (Sharma et al. 2015). Lower cytotoxicity of biosurfactant is expected for clinical applications.
Analytical characterization
FTIR analysis
The functional groups present in the biosurfactant produced by B. aryabhattai strain ZDY2 were examined by FTIR analysis to characterize the biosurfactant type. The FTIR spectrum is shown in Fig. 6a. A broad band at 3415.16 cm−1 represents the O–H stretching vibrations from free hydroxyl groups (Joy et al. 2017). A stretch around 3005.07 cm−1 corresponds to the presence of –CH3 and –CH2 groups in aliphatic chains of lipids (Elazzazy et al. 2015). Small signals at 2360.43 cm−1 may be due to the occurrence of R2C=N=N stretch (Barakat et al. 2017). Peak near 1788.67 cm−1 may be attributed to ester carbonyl groups such as –C = O bond of –COOH (Liu et al. 2017). A prominent peak at 1642.41 cm−1 was an indicative of existence of a peptide group in the molecule due to the characteristic amide vibrations (Barakat et al. 2017). Peak around 1384.29 cm−1 may be due to occurrence of C–O stretch of ester group (Chebbi et al. 2017). A broad band around 1147.67 cm−1 indicates a stretch of C–O band that occurs in lactones (El-Sheshtawy et al. 2016). Regions around 974.77 cm−1 signify alcohols and phenols (C–O–H) (Ibrahim et al. 2013). Peak near 885.80 cm−1 may be due to the vibrations of methylene scissoring of protein moieties present in the molecule (Elazzazy et al. 2015). Peak around 601.08 cm−1 may be probably due to the alkene of bacterial proteins (Barakat et al. 2017). Peak in the region of 502.48 cm−1 may be likely due to the presence of disulfides in the molecule (Ibrahim et al. 2013). The FTIR analysis reveals the lipopeptide nature of the biosurfactant produced by the bacteria. Several studies have reported the production of lipopeptide type of biosurfactants from different bacterial strains (Elazzazy et al. 2015; Bezza and Chirwa 2016). Similar spectrum of lipopeptide produced from Ochrobactrum and Bacillus genera was reported by Joy et al. (2017). Similarly, Barakat et al. (2017) showed the lipopeptide structure of biosurfactant from B. amyloliquefaciens SH20.
SEM–EDX analysis
The SEM image of biosurfactant produced by B. aryabhattai strain ZDY2 is shown in Fig. 6b. The image shows the surface of the biosurfactant which is polymeric in nature. The adherence of microbial cell to the polymeric molecule is evident. The biosurfactant image was captured on fourth day of biosurfactant production. The biosurfactant production was progressively induced by crude oil (hydrocarbon). Previous reports on SEM analysis to study the production of biosurfactant and cell adherence are available (Jain et al. 2013; Ohadi et al. 2017). The qualitative elemental analysis by SEM–EDS is shown in Fig. 6c. The elemental analysis reveals the mass and atomic percentage of elements such as carbon and oxygen that are largely present in the biosurfactant representing the carbonyl functional groups. The presence of such functional groups was also studied previously by Jain et al. (2012).
Conclusions
In the present research, biosurfactant was produced from an isolated B. aryabhattai strain ZDY2. The biosurfactant production kinetics revealed crude oil as inducing substrate and biosurfactant to be growth associated. The biosurfactant production was statistically optimized by CCD-RSM design and an increase of 2.51-fold. The interactions among crude oil–yeast extract and crude oil–NaNO3 were significant. The biosurfactant demonstrated thermal, pH and salinity stability. Strong antimicrobial potential of biosurfactant was revealed against several pathogens. Tested biosurfactant revealed lower cytotoxicity suggesting the safety of extracted biosurfactant. Lipopeptide nature of biosurfactant was revealed by FTIR spectroscopy. SEM–EDX analysis revealed the polymeric nature which was progressively induced by crude oil (hydrocarbon). The study interestingly suggests the antimicrobial biosurfactant from the isolated strain. Reports on lipopeptide type of biosurfactant production and optimization from B. aryabhattai strain are scare.
References
Almansoory AF, Hasan HA, Idris M, Abdullah SRS, Anuar N, Tibin EMM (2017) Biosurfactant production by the hydrocarbon-degrading bacteria (HDB) Serratia marcescens: optimization using central composite design (CCD). J Ind Eng Chem 47:272–280
Astuti DI, Purwasena IA, Putri RE, Amaniyah M, Sugai Y (2019) Screening and characterization of biosurfactant produced by Pseudoxanthomonas sp. G3 and its applicability for enhanced oil recovery. J Petrol Explor Prod Technol 9:2279–2289
Bagewadi ZK, Mulla SI, Shouche Y, Ninnekar HZ (2016) Xylanase production from Penicillium citrinum isolate HZN13 using response surface methodology and characterization of immobilized xylanase on glutaraldehyde-activated calcium-alginate beads. 3 Biotech 6:164
Bagewadi ZK, Mulla SI, Ninnekar HZ (2017) Optimization of laccase production and its application in delignification of biomass. Int J Recycl Org Waste Agric 6:351–365
Bagewadi ZK, Mulla SI, Ninnekar HZ (2018) Response surface methodology based optimization of keratinase production from Trichoderma harzianum isolate HZN12 using chicken feather waste and its application in dehairing of hide. J Environ Chem Eng 6:4828–4839
Bagewadi ZK, Muddapur UM, Madiwal SS, Mulla SI, Khan A (2019) Biochemical and enzyme inhibitory attributes of methanolic leaf extract of Datura inoxia Mill. Environ Sustain 2:75–87
Banat IM, Satpute SK, Cameotra SS, Patil Nyayanit NV (2014) Cost effective technologies and renewable substrates for biosurfactants’ production. Front Microbiol 5:1–18
Barakat KM, Hassan SWM, Darwesh OM (2017) Biosurfactant production by haloalkaliphilic Bacillus strains isolated from Red Sea, Egypt. Egypt J Aquat Res 43:205–211
Bezza FA, Chirwa EMN (2016) Biosurfactant-enhanced bioremediation of aged polycyclic aromatic hydrocarbons (PAHs) in creosote contaminated soil. Chemosphere 144:635–644
Bhat SS, Revankar VK, Kumbar V, Bhat K, Kawade VA (2018) Synthesis, crystal structure and biological properties of a cis-dichloridobis (diimine) copper(II) complex. Acta Cryst 74:146–151
Chebbi A, Hentati D, Zaghden H, Baccar N, Rezgui F, Chalbi M, Sayadi S, Chamkha M (2017) Polycyclic aromatic hydrocarbon degradation and biosurfactant production by a newly isolated Pseudomonas sp. strain from used motor oil-contaminated soil. Int Biodeter Biodegr 122:128–140
Ciandrini E, Campana R, Casettari L, Perinelli DR, Fagioli L, Manti A, Palmieri GF, Papa S, Baffone W (2016) Characterization of biosurfactants produced by Lactobacillus spp. and their activity against oral streptococci biofilm. Appl Microbiol Biotechnol 100:6767–6777
Deepika KV, Kalam S, Sridhar PR, Podile AR, Bramhachari PV (2016) Optimization of rhamnolipid biosurfactant production by mangrove sediment bacterium Pseudomonas aeruginosa KVD-HR42 using response surface methodology. Biocatal Agric Biotechnol 5:38–47
Derguine-Mecheri L, Kebbouche-Gana S, Khemili-Talbi S, Djenane D (2018) Screening and biosurfactant/bioemulsifier production from a high-salt-tolerant halophilic Cryptococcus strain YLF isolated from crude oil. J Petrol Sci Eng 162:712–724
Dwivedia A, Kumara A, Bhat JL (2019) Production and characterization of biosurfactant from Corynebacterium species and its effect on the growth of petroleum degrading bacteria. Microbiology 88(1):87–93
Elazzazy AM, Abdelmoneim TS, Almaghrabi OA (2015) Isolation and characterization of biosurfactant production under extreme environmental conditions by alkali-halo-thermophilic bacteria from Saudi Arabia. Saudi J Biol Sci 22:466–475
El-Sheshtawy HS, Aiad I, Osman ME, Abo-ELnasr AA, Kobisy AS (2016) Production of biosurfactants by Bacillus licheniformis and Candida albicans for application in microbial enhanced oil recovery. Egypt J Pet 25:293–298
Elshikh M, Marchant R, Banat IM (2016) Biosurfactants: promising bioactive molecules for oral-related health applications. FEMS Microbiol Lett 363(18):213
Fai AEC, Simiqueli APR, Andrade CJ, Ghiselli G, Pastore GM (2015) Optimized production of biosurfactant from Pseudozyma tsukubaensis using cassava wastewater and consecutive production of galactooligosaccharides: an integrated process. Biocatal Agric Biotechnol 4:535–542
Geys R, Soetaert W, Bogaert IV (2014) Biotechnological opportunities in biosurfactant production. Curr Opin Biotechnol 30:66–72
Hemlata B, Selvin J, Tukaram K (2015) Optimization of iron chelating biosurfactant production by Stenotrophomonas maltophilia NBS-11. Biocatal Agric Biotechnol 4:135–143
Heryani H, Putra MD (2017) Kinetic study and modeling of biosurfactant production using Bacillus sp. Electron J Biotechnol 27:49–54
Ibrahim ML, Ijah UJJ, Manga SB, Bilbis LS, Umar S (2013) Production and partial characterization of biosurfactant produced by crude oil degrading bacteria. Int Biodeter Biodegr 81:28–34
Ismail W, Shammary SAL, El-Sayed WS, Obuekwe C, El Nayal AM, Raheem ASA, Al-Humam A (2015) Stimulation of rhamnolipid biosurfactants production in Pseudomonas aeruginosa AK6U by organosulfur compounds provided as sulfur sources. Biotechnol Rep 7:55–63
Jain RM, Mody K, Mishra A, Jha B (2012) Isolation and structural characterization of biosurfactant produced by an alkaliphilic bacterium Cronobacter sakazakii isolated from oil contaminated wastewater. Carbohyd Polym 87:2320–2326
Jain RM, Mody K, Joshi N, Mishra A, Jha B (2013) Effect of unconventional carbon sources on biosurfactant production and its application in bioremediation. Int Biol Macromol 62:52–58
Joy S, Rahman PKSM, Sharma S (2017) Biosurfactant production and concomitant hydrocarbon degradation potentials of bacteria isolated from extreme and hydrocarbon contaminated environments. Chem Eng J 317:232–241
Khopade A, Biao R, Liu X, Mahadik K, Zhang L, Kokare C (2012) Production and stability studies of the biosurfactant isolated from marine Nocardiopsis sp. B4. Desalination 285:198–204
Kumar AP, Janardhan A, Radha S, Viswanath B, Narasimha G (2015) Statistical approach to optimize production of biosurfactant by Pseudomonas aeruginosa 2297. 3 Biotech 5:71–79
Kumari B, Singh SN, Singh DP (2012) Characterization of two biosurfactant producing strains in crude oil degradation. Process Biochem 47:2463–2471
Liu C, You Y, Zhao R, Sun D, Zhang P, Jiang J, Zhu A, Liu W (2017) Biosurfactant production from Pseudomonas taiwanensis L1011 and its application in accelerating the chemical and biological decolorization of azo dyes. Ecotox Environ Safe 145:8–15
Makkar RS, Cameotra SS, Banat IM (2011) Advances in utilization of renewable substrates for biosurfactant production. AMB Exp 1:5
Morais IMC, Cordeiro AL, Teixeira GS, Domingues VS, Nardi RMD, Monteiro AS, Alves RJ, Siqueira EP, Santos VL (2017) Biological and physicochemical properties of biosurfactants produced by Lactobacillus jensenii P6A and Lactobacillus gasseri P65. Microb Cell Fact 16:155
Mouafi FE, Elsoud MMA, Moharam ME (2016) Optimization of biosurfactant production by Bacillus brevis using response surface methodology. Biotechnol Rep 9:31–37
Mulla SI, Sun Q, Hu A, Wang Y, Ashfaq M, Eqani SAMAS, Yu CP (2016) Evaluation of sulfadiazine degradation in three newly isolated pure bacterial cultures. PLoS ONE 11(10):e0165013
Mulla SI, Hu A, Sun Q, Li J, Suanon F, Ashfaq M, Yu CP (2018) Biodegradation of sulfamethoxazole in bacteria from three different origins. J Environ Manag 206:93–102
Nalini S, Parthasarathi R (2018) Optimization of rhamnolipid biosurfactant production from Serratia rubidaea SNAU02 under solid-state fermentation and its biocontrol efficacy against Fusarium wilt of eggplant. Ann Agrar Sci 16:108–115
Nawawi WMFW, Jamal P, Alam MZ (2010) Utilization of sludge palm oil as a novel substrate for biosurfactant production. Bioresour Technol 101:9241–9247
Ndlovu T, Rautenbach M, Vosloo JA, Khan S, Khan W (2017) Characterisation and antimicrobial activity of biosurfactant extracts produced by Bacillus amyloliquefaciens and Pseudomonas aeruginosa isolated from a wastewater treatment plant. AMB Exp 7:108
Ohadi M, Dehghannoudeh G, Shakibaie M, Banat IM, Pournamdari M, Forootanfar H (2017) Isolation, characterization, and optimization of biosurfactant production by an oil-degrading Acinetobacter junii B6 isolated from an Iranian oil excavation site. Biocatal Agric Biotechnol 12:1–9
Paraszkiewicz K, Bernat P, Kusmierska A, Chojniak J, Płaza G (2018) Structural identification of lipopeptide biosurfactants produced by Bacillus subtilis strains grown on the media obtained from renewable natural resources. J Environ Manage 209:65–70
Plackett RL, Burman JP (1946) The design of optimum multifactorial experiments. Biometrika 33:305–325
Pruthi V, Cameotra SS (1997) Production of a biosurfactant exhibiting excellent emulsification and surface active properties by Serratia marcescens. World J Microbiol Biotechnol 13:133–135
Sachdev DP, Cameotra SS (2013) Biosurfactants in agriculture. Appl Microbiol Biotechnol 97:1005–1016
Sambanthamoorthy K, Feng X, Patel R, Patel S, Paranavitana C (2014) Antimicrobial and antibiofilm potential of biosurfactants isolated from lactobacilli against multi-drug-resistant pathogens. BMC Microbiol 14:197
Santos DKF, Brandao YB, RufinoRD Luna JM, Salgueiro AA, Santos VA, Sarubbo LA (2014) Optimization of cultural conditions for biosurfactant production from Candida lipolytica. Biocatal Agric Biotechnol 3:48–57
Santos DKF, Meira HM, Rufino RD, Luna JM, Sarubbo LA (2017) Biosurfactant production from Candida lipolytica in bioreactor and evaluation of its toxicity for application as a bioremediation agent. Process Biochem 54:20–27
Sharma D, Saharan BS, Chauhan N, Procha S, Lal S (2015) Isolation and functional characterization of novel biosurfactant produced by Enterococcus faecium. SpringerPlus 4:4
Shekhar S, Sundaramanickam A, Balasubramanian T (2014) Biosurfactant producing microbes and their potential applications: a review. Crit Rev Environ Sci Technol 45:1522–1554
Souza KST, Gudina EJ, Schwan RF, Rodrigues LR, Dias DR, Teixeira JA (2018) Improvement of biosurfactant production by Wickerhamomyces anomalus CCMA 0358 and its potential application in bioremediation. J Hazard Mater 346:152–158
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Varjani SJ, Upasani VN (2017) Critical review on biosurfactant analysis, purification and characterization using rhamnolipid as a model biosurfactant. Bioresour Technol 232:389–397
Acknowledgements
The first author and the corresponding author thank KLE Technological University, Hubballi, for providing funding to carry out the present research through capacity building projects.
Author information
Authors and Affiliations
Contributions
DY and ZB have designed the experimental work and executed the experiments. DY, ZB, UM and SM have carried our data analysis. DY and ZB have written the manuscript. All the authors have approved the contents of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
All the authors affirm that they have no argument of interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yaraguppi, D.A., Bagewadi, Z.K., Muddapur, U.M. et al. Response surface methodology-based optimization of biosurfactant production from isolated Bacillus aryabhattai strain ZDY2. J Petrol Explor Prod Technol 10, 2483–2498 (2020). https://doi.org/10.1007/s13202-020-00866-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13202-020-00866-9