Abstract
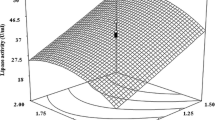
Enterococcus durans NCIM5427 (ED-27), capable of producing an intracellular acid stable lipase, was isolated from fish processing waste. Its growth and subsequent lipase production was optimized by Box Behneken design (optimized conditions: 5 % v/v fish waste oil (FWO), 0.10 mg/ml fish waste protein hydrolysates (FWPH) at 48 h of fermentation time). Under optimized conditions, ED-27 showed a 3.0 fold increase (207.6 U/ml to 612.53 U/ml) in lipase production, as compared to un-optimized conditions. Cell growth and lipase production was modeled using Logistic and Luedeking-Piret model, respectively; and lipase production by ED-27 was found to be growth-associated. Lipase produced by ED-27 showed stability at low pH ranges from 2 to 5 with its optimal activity at 30 °C , pH 4.6; showed metal ion dependent activity wherein its catalytic activity was activated by barium, sodium, lithium and potassium (10 mM); reduced by calcium and magnesium (10 mM). However, iron and mercury (5 mM) completely inactivated the enzyme. In addition, modifying agents like SDS, DTT, β-ME (1%v/v) increased activity of lipase of ED-27; while, PMSF, DEPC and ascorbic acid resulted in a marked decrease. ED-27 had maximum cell growth of 9.90309 log CFU/ml under optimized conditions as compared to 13 log CFU/ml in MRS. The lipase produced has potential application in poultry and slaughterhouse waste management.





Similar content being viewed by others
References
Amit KR, Swapna HC, Bhaskar N, Halami PM, Sachindra NM (2010) Effect of fermentation ensilaging on recovery of oil from freshwater fish viscera. Enzyme Microb Technol 46:9–13
Bhaskar N, Benila T, Radha C, Lalitha RG (2008) Optimization of enzymatic hydrolysis of visceral waste proteins of Catla (Catla catla) for preparing protein hydrolysate using a commercial protease. Biores Technol 99:335–343
Borkar PS, Bodade RG, Rao SR, Khobragade CN (2009) Purification and characterization of extracellular lipase from a new strain Pseudomonas aeruginosa SRT9. Braz J Microbiol 40:358–366
Chouhan M, Dawande AY (2010) Partial purification, characterization of lipase produced from P. aeruginosa. Asiatic J Biotechnol Res 1:29–34
Dheeman DS, Frias JM, Henehan GTM (2010) Influence of cultivation conditions on the production of a thermostable extracellular lipase from Amycolatopsis mediterranei DSM 43304. J Ind Microbiol Biotechnol 37:1–17
Franz CMAP, Holzapfel WH, Stiles ME (1999) Enterococci at the crossroads of food safety. Int J Food Microbiol 47:1–24
Gombert AK, Nielson J (2000) Mathematical modeling of metabolism. Curr Opin Biotechnol 11:180–186
Gornall AG, Bardawill CS, David MM (1949) Determination of serum protein by means of Biuret reaction. J Biol Chem 177:751–766
Gupta R, Gupta N, Rathi P (2004) Bacterial lipases: an overview of production, purification and biochemical properties. Appl Microbiol Biotechnol 64:763–781
Gupta N, Sahai V, Gupta R (2007) Alkaline lipase from a novel strain Burkholderia multivorans: statistical medium optimization and production in a bioreactor. Process Biochem 42:518–526
Hasan F, Shah AA, Hameed A (2006) Industrial applications of microbial lipases. Enzyme Microb Technol 39:235–251
Horn SJ, Aspmo SI, Eijsink VGH (2005) Growth of Lactobacillus plantarum in media containing hydrolysates of fish viscera. J Appl Microbiol 99:1082–1089
Hou CT, Johnston TM (1992) Screening of lipase activity with cultures from Agricultural Research Service Culture Collection. J Am Oil Chem Soc 69:1088–1097
Kaur P, Satyanarayana T (2005) Production of cell-bound phytase by Pichia anomala in an economical cane molasses medium: optimization using statistical tools. Process Biochem 40:3095–3102
Kouker G, Jaeger KE (1987) Specific and sensitive plate assay for bacterial lipases. Appl Environ Microbiol 53:211–213
Kumari A, Mahapatra P, Banerjee R (2009) Statistical optimization of culture conditions by response surface methodology for synthesis of lipase with Enterobacter aerogenes. Braz Arch Biol Technol 52:1349–1356
Liu C, Lu W, Chand J (2006) Optimizing lipase production of Burkholderia sp. by response surface methodology. Process Biochem 41:1940–1944
Lopes MFS, Crespo MTB (1999) Influence of environmental factors on lipase production by Lactobacillus plantarum. Appl Microbiol Biotechnol 51:249–254
Luedeking R, Piret EL (1959) A kinetic study of the lactic acid fermentation: batch process at controlled pH. J Biochem Microbiol 1:393–431
Moreno MRF, Sarantinopoulos P, Tsakalidou E, De Vuyst L (2006) The role and application of enterococci in food and health. Int J Food Microbiol 106:1–24
Neves-Petersen MT, Fojan P, Petersen SB (2001) How do lipases and esterases work: the electrostatic contribution. J Biotechnol 85:115–147
Oh S, Sungsue Rheem S, Jaehun Sim J, Kim S (1995) Optimizing conditions for the growth of Lactobacillus casei YIT 9018 in tryptone-yeast extract-glucose medium by using response surface methodology. Appl Environ Microbiol 61:3809–3814
Rajendran A, Thangavelu V (2007) Sequential optimization of culture medium composition for extracellular lipase production by Bacillus sphaericus using statistical methods. J Chem Technol Biotechnol 82:460–470
Ramani K, Chockalingam E, Sekaran G (2010) Production of a novel extracellular acidic lipase from Pseudomonas gessardii using slaughterhouse waste as substrate. J Ind Microbiol Biotechnol 37(5):531–535
Rebah FB, Frikha F, Kamoun W, Belbahri L, Gargouri Y, Miled N (2008) Culture of Staphylococcus xylosus in fish processing by-product-based media for lipase production. Lett Appl Microbiol 47:545–554
Sarantinopoulos P, Andrighetto C, Georgalaki M, Lombardi A, Cogan TM, Georgalaki M, Tsakalidou E (2001) Biochemical properties of enterococci relevant to their technological performance. Int Dairy J 11:621–647
Sharma D, Sharma B, Shukla AK (2011) Biotechnological approach of microbial lipase: a review. Biotechnology 10:23–40
Shuhaimi M, Kabeir BM, Yazid AM, Somchit MN (2009) Synbiotics growth optimization of Bifidobacterium pseudocatenulatum G4 with prebiotics using a statistical methodology. J Appl Microbiol 106:191–198
Souissi N, Bougatef A, Triki-Ellouz Y, Nasri M (2009) Production of lipase and biomass by Staphylococcus simulans grown on Sardinella (Sardinella aurita) hydrolysates and peptone. Afr J Biotechnol 8:451–457
Statsoft (1999) Statistics for Windows, Statsoft Inc., Tulsa, USA
Teng Y, Xu Y (2008) Culture condition improvement for whole-cell lipase production in submerged fermentation by Rhizopus chinensis using statistical method. Biores Technol 99:3900–3907
Thapa N, Pal J, Tamang JP (2006) Phenotypic identification and technological properties of lactic acid bacteria isolated from traditionally processed fish products of the Eastern Himalayas. Int J Food Microbiol 107:33–38
Tsakalidou E, Manolopoulou E, Tsilibari V, Georgalaki M, Kalantzopoulous G (1993) Esterolytic activities of Enterococcus durans and E. faecium strains isolated from greek cheese. Neth Milk Dairy J 47:145–150
Vrinda R, Bijinu B, Amit KR, Halami PM, Bhaskar N (2012) Concomitant production of lipase, protease and enterocin by Enterococcus faecium NCIM5363 and Enterococcus durans NCIM 5427 isolated from fish processing waste. Int Aquatic Res 4:14. doi:10.1186/2008-6970-4-14
Wang Y, Zhao J, Xu J, Fan L, Li S, Zhao L, Mao X (2010) Significantly improved expression and biochemical properties of recombinant Serratia marcescens lipase as robust biocatalyst for kinetic resolution of chiral ester. Appl Biochem Biotechnol 162:2387–2399
Acknowledgments
BN thanks CSIR for funding this research work under the EMPOWER scheme (OLP 90). VR acknowledges the University Grants Commission (UGC) for the CSIR-UGC fellowship. Authors place on record their thanks to Director, CFTRI for permission to publish the work.
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Ramakrishnan, V., Goveas, L.C., Halami, P.M. et al. Kinetic modeling, production and characterization of an acidic lipase produced by Enterococcus durans NCIM5427 from fish waste. J Food Sci Technol 52, 1328–1338 (2015). https://doi.org/10.1007/s13197-013-1141-5
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13197-013-1141-5