Abstract
Purpose
Early deep-learning-based image denoising techniques mainly focused on a fully supervised model that learns how to generate a clean image from the noisy input (noise2clean: N2C). The aim of this study is to explore the feasibility of the self-supervised methods (noise2noise: N2N and noiser2noise: Nr2N) for PET image denoising based on the measured PET data sets by comparing their performance with the conventional N2C model.
Methods
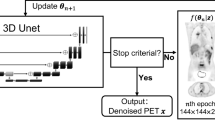
For training and evaluating the networks, 18F-FDG brain PET/CT scan data of 14 patients was retrospectively used (10 for training and 4 for testing). From the 60-min list-mode data, we generated a total of 100 data bins with 10-s duration. We also generated 40-s-long data by adding four non-overlapping 10-s bins and 300-s-long reference data by adding all list-mode data. We employed U-Net that is widely used for various tasks in biomedical imaging to train and test proposed denoising models.
Results
All the N2C, N2N, and Nr2N were effective for improving the noisy inputs. While N2N showed equivalent PSNR to the N2C in all the noise levels, Nr2N yielded higher SSIM than N2N. N2N yielded denoised images similar to reference image with Gaussian filtering regardless of input noise level. Image contrast was better in the N2N results.
Conclusion
The self-supervised denoising method will be useful for reducing the PET scan time or radiation dose.





Similar content being viewed by others
References
Lehtinen J, Munkberg J, Hasselgren J, Laine S, Karras T, Aittala M, et al. Noise2Noise: learning image restoration without clean data. arXiv. 2018;1803.04189.
Moran N, Schmidt D, Zhong Y, Coady P. Noisier2Noise: learning to denoise from unpaired noisy data. arXiv. 2019;1910.11908.
Teymurazyan A, Riauka T, Jans H-S, Robinson D. Properties of noise in positron emission tomography images reconstructed with filtered-backprojection and row-action maximum likelihood algorithm. J Digit Imaging. 2013;26:447–56.
Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. Pro Med Image Comput Comput Assist Interv. 2015:234–41.
Hegazy MAA, Cho MH, Cho MH, Lee SY. U-net based metal segmentation on projection domain for metal artifact reduction in dental CT. Biomed Eng Lett. 2018;9:375–85.
Hwang D, Kang SK, Kim KY, Seo S, Paeng JC, Lee DS, et al. Generation of PET attenuation map for whole-body time-of-flight 18F-FDG PET/MRI using a deep neural network trained with simultaneously reconstructed activity and attenuation maps. J Nucl Med. 2019;60:1183–9.
Lee MS, Hwang D, Kim HJ, Lee JS. Deep-dose: a voxel dose estimation method using deep convolutional neural network for personalized internal dosimetry. Sci Rep. 2019;9:10308.
Park J, Bae S, Seo S, Park S, Bang J-I, Han JH, et al. Measurement of glomerular filtration rate using quantitative SPECT/CT and deep-learning-based kidney segmentation. Sci Rep. 2019;9:4223.
Hwang D, Kim KY, Kang SK, Seo S, Paeng JC, Lee DS, et al. Improving the accuracy of simultaneously reconstructed activity and attenuation maps using deep learning. J Nucl Med. 2018;59:1624–9.
Lee JS. A review of deep learning-based approaches for attenuation correction in positron emission tomography. IEEE Trans Radiat Plasma Med Sci. 2020. https://doi.org/10.1109/TRPMS.2020.3009269.
Wang Z, Bovik A, Sheikh H, Simoncelli E. Image quality assessment: from error visibility to structural similarity. IEEE Trans Image Process. 2004:600–12.
Wu D, Gong K, Kim K, Li X, Li Q. Consensus neural network for medical imaging denoising with only noisy training samples. Proc Med Image Comput Comput Assist Interv. 2019:741–9.
Funding
This work was supported by grants from National Research Foundation of Korea (NRF) funded by Korean Ministry of Science, ICT and Future Planning (grant no. NRF-2016R1A2B3014645).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Si Young Yie, Seung Kwan Kang, Donghwi Hwang, and Jae Sung Lee declare that there is no conflict of interest.
Ethical Approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. The retrospective use of the scan data was approved by the Institutional Review Board of our institute.
Informed Consent
Waiver of consent was approved by the Institutional Review Board of our institute.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Yie, S.Y., Kang, S.K., Hwang, D. et al. Self-supervised PET Denoising . Nucl Med Mol Imaging 54, 299–304 (2020). https://doi.org/10.1007/s13139-020-00667-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13139-020-00667-2