Abstract
The order Ceraceosorales (Ustilaginomycotina) currently includes the single genus Ceraceosorus, with one species, Ceraceosorus bombacis, parasitic on Bombax ceiba in India. The diversity, biogeography, evolution, and phylogenetic relationships of this order are still relatively unknown. Here, a second species of Ceraceosorus is described from West Africa as a novel species, Ceraceosorus africanus, infecting Bombax costatum in Benin, Ghana, and Togo. This species produces conspicuous fructifications, similar to corticioid basidiomata when mature, but sorus-like in early stages of ontogenetic development. The fructifications cover much of the leaf surface and resemble leaf blight. This contrasts with the inconspicuous fructifications of C. bombacis comprising small spots scattered over the lower leaf surface that resemble leaf spot. Both species of Ceraceosorus differ in several micromorphological traits, infect different host plant species in widely separated geographical areas, and are separated by a considerable genetic distance in 28S rDNA and RPB2 genes. The distinct corticioid fructification of C. africanus is a unique morphological trait within the Ustilaginomycotina. Molecular phylogenetic analyses of a single gene dataset (D1/D2 28S rDNA) supported the monophyly of the two Ceraceosorus species and the Ceraceosorales and their placement within the Ustilaginomycotina. Molecular phylogenetic analyses of a multigene dataset (18S/5.8S/28S rDNA/RPB2/TEF1) revealed Exobasidium rhododendri (Exobasidiales) as the closest relative of Ceraceosorus, both clustering together with Entyloma calendulae (Entylomatales), indicating affinities to the Exobasidiomycetes. This phylogenetic placement is in agreement with ultrastructural characteristics (presence of local interaction zone and interaction apparatus) reported for the Ceraceosorales, Entylomatales, and Exobasidiales.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Historically, smut fungi were defined as phytoparasites producing (usually) dusty masses of dikaryotic teliospores within plant tissues, germinating to develop basidia with basidiospores, which grow as a saprobic yeast stage in the haplophase. They were included in the single order Ustilaginales (Schröter 1889; Clinton 1906; Zundel 1953). Parasites with such a life strategy and organization are now referred to as classical smuts (Vánky 2001), true smuts (Oberwinkler 2012), or teliosporic smuts (Begerow et al. 2006). This idea of smut fungi and their classification remained almost unchanged until the end of the twentieth century. Comprehensive studies by Bauer et al. (1997) on hyphal septal pores and interaction zones between parasites and host cells using transmission electron microscopy revealed enormous structural diversity that led to the new classification and changed the concept of smut fungi and their relationships to other fungal parasites. These findings were supported by the molecular phylogenetic studies of Begerow et al. (1997). These two complementary studies demonstrated that the classical smut fungi (at that time assigned to the single order Ustilaginales) represented several distinct lineages regarded as the orders Doassansiales, Entorrhizales, Entylomatales, Georgefischeriales, Tilletiales, Urocystidales, and Ustilaginales. These orders were interspersed by nonteliosporic taxa forming two main lineages referred to as the orders Exobasidiales (incl. Graphiolales, Begerow et al. 2002a) and Microstromatales. The new concept of smuts and related nonteliosporic fungi resulted in the description of the new class Ustilaginomycetes (Bauer et al. 1997), that was subsequently elevated to the level of a subphylum, the Ustilaginomycotina (Bauer et al. 2006). It became apparent that several genera and species should be removed from the smut fungi since they showed relationships to rust fungi. These are now referred to as false smuts (Oberwinkler 2012) and are classified in the Pucciniomycotina, order Microbotryales (Bauer et al. 1997; Begerow et al. 1997). Recently, two additional genera, Entorrhiza C.A. Weber and Talbotiomyces Vánky, R. Bauer & Begerow, were removed from the smut fungi and classified in the new phylum, Entorrhizomycota, distinct from the phyla Ascomycota and Basidiomycota (Bauer et al. 2015; Riess et al. 2015).
The new concept of smuts and related fungi was continuously augmented by further phylogenetic studies that revealed an enormous diversity of life strategies and structural organization of sporulation within the lineages of Ustilaginomycotina. Thus, in addition to classical smut fungi and nonteliosporic species, this subphylum includes lipophilic yeasts associated with warm-blooded animals or marine environments, the species of Malassezia Baill. (Begerow et al. 2000; Amend 2014; Cabañes 2014; Wang et al. 2014), mite-associated yeasts (Boekhout et al. 2003), and saprobic yeasts isolated from diverse ecosystems (Begerow et al. 2000; Boekhout et al. 2006; Nasr et al. 2014; Piątek et al. 2015; Wang et al. 2014). Several species were also isolated from healthy plant tissues and may act as endophytes (Paz et al. 2007; Amin et al. 2010; Takahashi et al. 2011; Padhi and Tayung 2013; Rush and Aime 2013). The most notable of them is the recently described Violaceomyces palustris Albu, Toome & Aime that represents a distinct lineage, the order Violaceomycetales (Albu et al. 2015). Sporulation in the Ustilaginomycotina is notably diverse, including budding cells, simple hyphae with conidiogenous cells producing conidia, teliospores embedded between plant cells or replacing generative or vegetative plant organs, and loose fascicles or hymenial layers of basidia emerging from stomata or the epidermis.
To date Ceraceosorus B.K. Bakshi is a monotypic genus and contains only Ceraceosorus bombacis (B.K. Bakshi) B.K. Bakshi, a leaf pathogen of the silk cotton tree Bombax ceiba L. (Malvaceae) in Uttar Pradesh, India. This fungus was found and initially described as Dicellomyces bombacis B.K. Bakshi by Bakshi (in Bakshi et al. 1972). Only 3 years later, it was transferred, also by Bakshi, to the new genus Ceraceosorus, distinct from Dicellomyces L.S. Olive (Olive 1945, 1951) in having sorus-like fructifications, indeterminate hymenial thickenings, intracellular hyphae, basidia with internal thickenings, and lacking cystidia (Cunningham et al. 1976). Ceraceosorus was placed in the Brachybasidiaceae (Cunningham et al. 1976; Kirk et al. 2001). C. bombacis may be an important phytopathogenic species in India as reported in the original publications (Bakshi et al. 1972; Cunningham et al. 1976), though in recent years, it was not included as such in any original phytopathological report. C. bombacis is unique in respect to the formation of intracellular hyphae, a character not known in any other order of Exobasidiomycetes, stressing the isolated phylogenetic position of this species outside all main lineages revealed by molecular phylogenetic analyses (Begerow et al. 2006). Thus, to accommodate the genus Ceraceosorus, Begerow et al. (2006) added the novel order Ceraceosorales, assigned to the class Exobasidiomycetes. The family Ceraceosoraceae was subsequently described by Denchev and Moore (2009).
The single genus and species currently residing in the Ceraceosorales implies that different aspects such as evolution, biogeography, diversity, and phylogenetic relationships of this order are still obscurely known. Some of these missing aspects could be improved now from the discovery of a second member of Ceraceosorales infecting a native woody species, the silk cotton tree Bombax costatum Pellegr. & Vuillet (Malvaceae), in the Sudanian savanna biome in West Africa. This unknown fungus was consistently found on most specifically inspected B. costatum trees in Benin, Ghana, and Togo during four independent surveys at the end of the rainy period in the years 2011–2013 and 2015. In this study, polyphasic analyses were conducted to establish the systematic and phylogenetic placement of this fungus, studying its structural (dis-)similarity and genetic divergence compared to Ceraceosorus bombacis, as well as phylogenetic relationships to other orders of the Ustilaginomycotina. Furthermore, environmental information assembled in field studies is given to contribute to the knowledge of the ecology of this fungus.
Material and methods
Specimen sampling and documentation
This study is based on phylogenetic and morphological analyses of a leaf parasite on Bombax costatum (Malvaceae) originating from eight locations in three different countries and diverse habitats in West Africa (Benin, Ghana, Togo; Figs. 1 and 2). The voucher specimens were deposited in the fungal collection of the W. Szafer Institute of Botany of the Polish Academy of Sciences, Kraków, Poland (KRAM F). Additionally, the ex-type culture of Ceraceosorus bombacis isolated from Bombax ceiba (Cunningham et al. 1976) was anew sequenced for phylogenetic analyses. Detailed specimen information is given in Table 1. Nomenclatural novelty was registered in MycoBank (www.mycobank.org).
Currently known distribution of Ceraceosorus africanus (red dots) on the background of the global distribution of Bombax costatum in the Sudanian savanna biome (green area, based on the map shown on the website: http://www.hombori.org/francais/biodiversity/flora.html)
Habitat types of Ceraceosorus africanus: a Sudanian savanna dominated by grasses, with scattered trees, between Zibogo and Tugu in Ghana; b Sudanian savanna with shrubs and trees, between Busunu and Fufulsu in Ghana (type locality); c transition zone between gallery forest and the Sudanian woodland, Forêt Classée de Béléfoungou in Benin; d inselberg, Tébou in Benin
Morphological analyses
Morphological characters of fructifications were studied using dried herbarium material. Observations of the macroscopic appearance of fructifications were made with the naked eye and under a stereoscopic microscope Nikon SMZ-2T. Micromorphological characters were studied by light microscopy. For this purpose, thin freehand sections of fructifications were made under the stereoscopic microscope using a razor blade; mounted in 3 % (wt/vol) aqueous potassium hydroxide, 1 % (wt/vol) aqueous phloxine, Melzer’s reagent, and 0.1 % Cotton Blue (wt/vol) in 60 % (wt/vol) lactic acid; and examined under a Nikon Eclipse E-400 light microscope. In presenting the spore size variation, 5 % of measurements were excluded from each end of the range and are given in parentheses. In the species description, the following abbreviations were used: L = mean length of all spore measurements, W = mean spore width, Q = length to width range ratio, Q m = mean Q value, and n = number of measurements. Q values were obtained from dividing the average basidiospore length by width. Line drawings were made from slides mounted in aqueous potassium hydroxide.
Molecular phylogenetic analyses
For DNA isolation, approximately 0.25 cm2 of dried leaves of Bombax costatum with disease symptoms and a 0.25 cm2 agar block containing a living culture of Ceraceosorus bombacis, respectively, were selected, deep-frozen in liquid nitrogen, and ground several times with a plastic pestle. Total genomic DNA was subsequently extracted using the InnuPREP Plant DNA Kit (Analytik Jena, Jena, Germany) following the standard protocol. C. bombacis was processed again since no 5.8S, RPB2, or TEF1 sequences were available, and only a short 28S sequence was available in the NCBI’s GenBank database (www.ncbi.nlm.nih.gov).
The nuclear 18S, 5.8S, and 28S ribosomal DNA (rDNA) genes (=18S, 5.8S, 28S) were amplified using the primer combinations NS1/NS8 (White et al. 1990), ITS3/ITS4 (White et al. 1990), and LR0R/LR9 (R. Vilgalys lab, http://biology.duke.edu/fungi/mycolab/primers.htm; Hopple and Vilgalys 1999), respectively, with the PCR conditions described in Riess et al. (2013). The gene of the second largest subunit of RNA polymerase II (RPB2) regions 5–7 was amplified with the primers fRPB2-5F (Liu et al. 1999) and bRPB2-7.1R (Matheny 2005), following the thermocycling program described by Matheny (2005). The translation elongation factor 1-alpha (TEF1) gene was amplified using the primer combinations EF-526F/EF-2218R, EF-526F/EF-ir, or Ef-df/EF-2218R (Rehner and Buckley 2005; S. Rehner, http://aftol.org/pdfs/EF1primer.pdf) following the protocol of Rehner and Buckley (2005). PCR products were cleaned and cycle sequenced as described in Riess et al. (2013) using the PCR primers and the additional primers NS19 (Gargas and Taylor 1992) and NS4 (White et al. 1990) for 18S; LR3R (Hopple and Vilgalys 1999) and LR6 (Vilgalys and Hester 1990) for 28S; fRPP2-6F (Matheny 2005) for RPB2; and EF-983F, EF-1567R, and EF-1577F (Rehner and Buckley 2005) for TEF1. All generated Ceraceosorus DNA sequences have been deposited in NCBI’s GenBank database (www.ncbi.nlm.nih.gov) (see Tables 1 and 2).
To infer the phylogenetic relationships of the examined Ceraceosorus specimens, we assembled two datasets. Dataset 1 contained D1/D2 28S sequences of seven Ceraceosorus specimens and representatives of all smut genera belonging to the main lineages of Ustilaginomycotina (Begerow et al. 2014; Wang et al. 2014) for which sequences were available in GenBank, including 73 sequences obtained from the respective generic type species (Fig. 3). Dataset 2 was composed of 18S + 5.8S + 28S + RPB2 + TEF1 sequences of Ceraceosorus plus one species of all Ustilaginomycotina genera for which at least four of these five genes were available in GenBank (21 total; Fig. 4). Microbotryum violaceum (Pers.) G. Deml & Oberw. s.l. and Puccinia graminis Pers. were used as outgroup for both datasets. Except for Ceraceosorus bombacis, Entyloma calendulae (Oudem.) de Bary, Schizonella melanogramma (DC.) J. Schröt., Sympodiomycopsis paphiopedili Sugiy., Tokuoka & Komag., Urocystis colchici (Schltdl.) Rabenh., and Ustanciosporium standleyanum (Zundel) M. Piepenbr., parts of the concatenated sequences of the remaining species were obtained from different specimens/cultures of the species (for detailed information, compare GenBank accessions). For GenBank accession numbers of the sequences of dataset 1 (Boekhout et al. 1995, 2003; Begerow et al. 1997, 2000, 2001, 2002a, 2006; Bauer et al. 1999, 2001, 2005, 2007, 2008; Piepenbring et al. 1999, 2002, 2010; Fell et al. 2000; Castlebury et al. 2005; Hendrichs et al. 2005; Stoll et al. 2005; Maier et al. 2006; Matheny et al. 2006; Vánky et al. 2006, 2008, 2013; González et al. 2007; Chandra and Huff 2008; Lutz et al. 2008, 2012a, b; Paap et al. 2008; Ritschel et al. 2008; Tanaka et al. 2008; Sipiczki and Kajdacsi 2009; Deadman et al. 2011; Vánky and Lutz 2011; McTaggart et al. 2012; Piątek et al. 2013; Nasr et al. 2014), see Fig. 3. For GenBank accession numbers of the sequences of dataset 2 (de Wachter et al. 1992; Boekhout et al. 1995; Schillberg et al. 1995; Swann and Taylor 1995; Takashima and Nakase 1996; Bakkeren et al. 2000; Döring and Blanz 2000; Hamamoto et al. 2000; Döring 2003; Liu and Hall 2004; Lutzoni et al. 2004; Wingfield et al. 2004; Castlebury et al. 2005; Stoll et al. 2005; Begerow et al. 2006; de Beer et al. 2006; Kolařík et al. 2006; Matheny et al. 2006, 2007; Carris et al. 2007; Le Gac et al. 2007; Ran et al. 2008; Brock et al. 2009; Rosa et al. 2009; Kottke et al. 2010; Gorfer et al. 2011; Schoch et al. 2012; Wang et al. 2014), see Table 2.
Phylogenetic placement of Ceraceosorus africanus within the sampled Ustilaginomycotina derived from D1/D2 28S sequences (447 bp). Statistical support is given as ML bootstrap above branches (≥70) and Bayesian posterior MCMC probability below branches (≥0.90). The lines in bold indicate a maximum support of 100/1.00. Microbotryum violaceum and Puccinia graminis were used as outgroup
Hypothesis of phylogenetic relationships of the sampled Ustilaginomycotina based on 18S, 5.8S, 28S, RPB2, and TEF1 sequences (4896 bp). The tree was rooted with Microbotryum violaceum s.l. and Puccinia graminis. Statistical support is given as ML bootstrap above branches (≥70) and Bayesian posterior MCMC probability below branches (≥0.90). The lines in bold indicate a maximum support of 100/1.00. The superorder Exobasidianae is marked with a black dot. Mon. = Moniliellomycetes
All DNA regions were aligned separately with MAFFT 7.147b using the E-INS-i option (Katoh et al. 2005; Katoh and Standley 2013). In the rDNA datasets, ambiguously aligned regions were removed using Gblocks 0.91b (Castresana 2000) with gaps allowed at alignment positions at which gaps were present in more than half of the sequences, while the protein coding sequences were modified manually with the help of annotated sequences of RPB2 (DQ408132) and TEF1 (X73529). The final alignment length for dataset 1 (D1/D2 28S) was 447 bp (15 % of the initial alignment length before Gblocks), and for dataset 2 (18S + 5.8S + 28S + RPB2 + TEF1) 1505 bp for partial 18S (54 %), 163 bp for complete 5.8S, 1272 bp for partial 28S (48 %), 972 bp for the complete exon 6 of RPB2, and 984 bp for partial TEF1. The single-gene alignments were then concatenated into one sequence alignment of a total of 4896 bp (dataset 2). Phylogenetic analyses were computed for both datasets using maximum likelihood (ML) with combined rapid bootstrapping under the GTRCAT model from 1000 runs using RAxML 8.0.17 (Stamatakis 2014). Additional posterior probability nodal support values were determined in a Bayesian phylogenetic MCMC search using MrBayes 3.2.2 (Ronquist et al. 2012) under the general time reversible model with gamma-distributed rate variation (GTR + G) as suggested in the MrBayes version 3.2 manual. Each search comprised two runs of four chains each for 10 × 106 generations sampled every 100 generations with the first 2.5 × 106 generations discarded as burn-in. Genetic divergences (uncorrected p-distance) within the Ceraceosorales were calculated for 18S, 28S, and RPB2 using Mesquite 2.75 (Maddison and Maddison 2011) and the unmodified alignments.
Results
Morphology: ontogenetic development
The fungus on Bombax costatum developed intracellular hyphae in host cells that agglomerated, as intercellular hyphae, in substomatal cavities and emerged through stomata forming sorus-like fructifications with a densely packed and strongly agglutinated hyphal subhymenial layer (that could be referred to as sterile external stroma), and an outer hymenial layer with basidioles and basidia. The adjoining sorus-like fructifications coalesced to form a continuous, compact, widely effused layer on the host leaves. The single sorus-like fructifications were visible best at the edge of the continuous layer, whereas after coalescence, they were undifferentiated and whole fructifications resembled those of corticioid fungi. The predominant parts of the fructifications were sterile, composed only of basidioles, waxy and pinkish in color. The fertile areas were scattered over the sterile areas and were visible under lens as whitish and slightly pruinose areas. The detailed morphological characterization of the fungus on B. costatum is included in the species description and depicted in Figs. 5a–f, 6, 7, and 8.
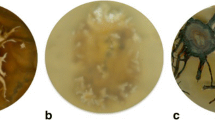
a–f Macroscopic symptoms of infection of Bombax costatum leaves by Ceraceosorus africanus (a–d taken on the type locality; e, f taken on the locality between Zibogo and Tugu): a view of the crown of the tree with scattered infected leaves marked by white arrows; b–f different levels of development of leaf blight caused by the fungus (b, c, e, f on the leaf underside; d on the leaf upperside). g–i Macroscopic symptoms of infection of Bombax ceiba leaves by Ceraceosorus bombacis: leaf spot caused by the fungus (g schematized drawing, reproduced from the figure presented in Bakshi et al. 1972; h–i photo taken from isotype KRAM F-58224, note sorus-like fructifications; scale bars: h, i = 3 mm)
Developmental stages of fructifications of Ceraceosorus africanus (all from holotype): a, b initial sorus-like fructifications visible in the marginal parts that coalesce to form a continuous, compact, widely effused layer on the host leaves; c schematized drawing of individual sorus-like fructification, note intracellular hyphae indicated by black arrows; d schematized drawing of the mode of coalescence of individual sorus-like fructifications to form compact fructification; e, f individual sorus-like fructification seen by LM with hyphae agglomerated in the substomatal cavities and emerged through the stomata to form strongly agglutinated subhymenial and hymenial layers. Scale bars: a, b = 1 mm, c–f = 100 μm
Molecular phylogenetic analyses
D1/D2 28S sequences of the six specimens of the leaf parasite on Bombax costatum were identical. Their phylogenetic relationships, analyzed with 90 sequences representing sampled genera of the Ustilaginomycotina (plus two outgroup species from Pucciniomycotina), are shown in Fig. 3. In all analyses, the specimens of the leaf parasite on B. costatum clustered together and as sister lineage of Ceraceosorus bombacis. The sequence divergence to C. bombacis was 0.20 % or 1 bp (18S), 4.03 % or 83 bp (28S), and 18.5 % or 196 bp (RPB2). The monophyly of the leaf parasite on B. costatum and C. bombacis was highly supported in both Bayesian and ML analyses. However, the phylogenetic position of the Ceraceosorales within the Exobasidiomycetes was not resolved, and the Exobasidiomycetes were not supported as monophyletic (Fig. 3).
The phylogenetic analyses of the concatenated 18S/5.8S/28S/RPB2/TEF1 dataset (Table 2) revealed the representative of the Exobasidiales, Exobasidium rhododendri (Fuckel) C.E. Cramer, as the closest relative of Ceraceosorus bombacis supported by 83 ML bootstrap and 0.89 posterior probability (Fig. 4). The Ceraceosorales, Exobasidiales, Entylomatales, and Doassansiales were monophyletic, however, without statistical support. In congruence with the topology derived from D1/D2 28S sequences, the monophyly of the Exobasidiomycetes was not supported.
Taxonomy
Ceraceosorus africanus Piątek, K. Riess, Karasiński & M. Lutz, sp. nov.
MycoBank no. MB 816865
Etymology: The name refers to Africa, corresponding to the occurrence of this fungus on this continent contrasting with its sister species which occurs in India.
Type: Ghana, Northern Region: between Busunu and Fufulsu, ca. 18 km W of Fufulsu, 09° 09′ 36″ N, 01° 25′ 47″ W, elev. ca. 120 m a.s.l., on Bombax costatum, 5 November 2012, leg. M. Piątek & N.S. Yorou [holotype KRAM F-57385; type sequences are available in GenBank: KP413055 (18S), KP413036 (28S), and KP413030 (RPB2)].
Description: Parasitic on Bombax costatum. Fructifications on the leaves, infection local, only single leaves infected. Fructifications hypophyllous, (100–)200–500 μm thick, resupinate, widely effused, closely adnate, waxy, cracked in old specimens, initially sorus-like and forming circular patches up to 0.5 mm in diam. (best visible in the marginal parts of the fructifications), later coalescent forming continuous, compact areas up to 6.5 cm long, covering a part of the hypophyllous surface, usually about 1/2 or 2/3 of the leaf blade, typically starting from the petiole, exceptionally from other parts of the leaf blade, rarely covering the whole hypophyllous area; the corresponding epiphyllous area clearly discolored, usually yellowish or brownish, finally blackish and necrotic; the fructifications sometimes appear on the upper side of leaves but then only in the form of thin, linear parallel rows on both sides of central nerves. Hymenial surface smooth, white and slightly pruinose (visible under lens) in fertile parts, pinkish to pinkish brown in sterile parts. Hyphal system monomitic. Hyphae 1–3 μm wide, lacking clamps, thin- to thick-walled, mostly hyaline or pale yellow in substomatal cavities, sometimes with very fine, scattered oil drops inside; hyphae sparsely branched, straight or slightly sinuous, at first intracellular, later agglomerating, as intercellular hyphae, in substomatal cavities and emerging through the stomata to form a dense, compact structure in all parts of fructifications, including subhymenium. Hymenium forming a dense palisade of basidioles and basidia. Basidioles 3–4 μm wide, cylindrical, usually thin-walled, sometimes thick-walled with even walls up to 0.5 μm thick, often with granular oil drops inside. Paraphysoid hyphae sometimes present among basidioles, 0.4–0.8 μm wide, thin-walled, sparsely branched or unbranched. Basidia 48–82 × 3–5 μm (measured without sterigmata), dacrymycetoid, thin-walled, with cylindrical distal end, tapering toward basal septa, sometimes anastomozing in basal parts (below basal septa, H-shaped in outline), with two prominent sterigmata up to 15 μm long; sterigmata emerge from the apical part of basidioles, at first as semicircular, later as elongated and obtuse, and finally as subulate (pointed) projections, collapsed after releasing the basidiospores, and difficult to be found in old hymenia. Basidiospores (6.5–)9–16(−18) × (3–)3.5–5(−5.2) μm (L = 12.72 μm, W = 3.95 μm, Q = 2.16–4.33, Q m = 3.14, n = 90), elongated, lacrymoid, usually with curved apical part, obtuse at distal end, thin-walled, hyaline, with somewhat darkened and refractive apical part, inamyloid, acyanophilous. Conidiophores and conidia not observed.
Additional specimens examined (paratypes): Benin, Atakora Department: near the Tanougou Waterfalls (Chutes de Tanougou), ca. 55 km N of Natitingou, 10° 48′ 21″ N, 01° 26′ 16″ E, elev. ca. 265 m a.s.l., on Bombax costatum, 26 October 2011, leg. M. Piątek & N.S. Yorou (KRAM F-57386); Benin, Borgou Department: Bassa, ca. 12.5 km E of Parakou, 09° 17′ 08″ N, 02° 43′ 41″ E, elev. ca. 317 m a.s.l., on B. costatum, 28 October 2013, leg. M. Piątek (KRAM F-57390); Benin, Donga Department: Tébou, inselberg, ca. 28 km NE of Djougou, 09° 54′ 49″ N, 01° 48′ 36″ E, elev. ca. 485 m a.s.l., on B. costatum, 19 September 2015, leg. M. Piątek (KRAM F-58019); Benin, Donga Department: Forêt Classée de Béléfoungou, ca. 12 km NE of Djougou, 09° 47′ 54″ N, 01° 42′ 52″ E, elev. ca. 405 m a.s.l., on B. costatum, 19 September 2015, leg. M. Piątek (KRAM F-58020); Ghana, Northern Region: between Zibogo and Tugu, ca. 14 km E of Tamale, 09° 22′ 25″ N, 00° 43′ 04″ W, elev. ca. 130 m a.s.l., on B. costatum, 3 November 2012, leg. M. Piątek & N.S. Yorou (KRAM F-57387); Ghana, Northern Region: between Busunu and Fufulsu, ca. 13.5 km W of Fufulsu, 09° 09′ 05″ N, 01° 23′ 33″ W, elev. ca. 120 m a.s.l., on B. costatum, 5 November 2012, leg. M. Piątek & N.S. Yorou (KRAM F-57388); Togo, Kara Region: between Pya and Niamtougou, ca. 19 km N of Kara, 09° 42′ 10″ N, 01° 07′ 30″ E, elev. ca 345 m a.s.l., on B. costatum, 3 November 2013, leg. M. Piątek & N.S. Yorou (KRAM F-57389).
Comments: The description above is based on the holotype specimen from Ghana. The materials from other localities were roughly similar, with the exception of the specimen from Togo (KRAM F-57389) that differs in having 1-sterigmate basidia and longer basidiospores, which were up to 25 μm long. Such morphology of basidia and size of basidiospores are considered an abnormality or variation rather than the typical morphology of Ceraceosorus africanus. Basidiospores in 1-sterigmate basidia may be larger than in 2-sterigmate basidia because there are more nutrients available. Similar abnormalities in basidiospore sizes produced on 1–2-sterigmate and 4-sterigmate basidia were already observed in other fungi (Lebel and Castellano 2002; Kautmanová et al. 2012). The specimens collected in September 2015 from Benin were not fully mature.
Distribution and ecology: Ceraceosorus africanus was found at eight locations in Benin, Ghana, and Togo, within the Sudanian savanna biome. The northernmost location was found near the Tanougou Waterfalls in Benin, at about 10° 48′ 21″ N; the southernmost location ca. 13.5 km W of Fufulsu in Ghana, at about 09° 09′ 05″ N; the westernmost location ca. 18 km W of Fufulsu in Ghana, at about 01° 25′ 47″ W; and the easternmost location near Bassa in Benin, at about 02° 43′ 41″ E. These localities are situated in the core geographical range of Bombax costatum (Fig. 1), but it is likely that C. africanus occurs with the host plant over its distribution in West Africa. The habitat conditions for five of the eight locations where C. africanus was found were similar: open Sudanian savanna with scattered shrubs and trees, with infected trees of B. costatum fully exposed to the sun. At the locality near the Tanougou Waterfalls, the infected tree was found in the transition zone between gallery forest and rocky savanna in a semishaded place, at the locality in the Forêt Classée de Béléfoungou in the transition zone between gallery forest and the Sudanian woodland, and at the locality in Tébou on the top of the inselberg (Fig. 2). The first infection symptoms with immature fructifications were observed at the end of September (observations in 2015 in Benin). Infected leaves with mature fructifications were found at the end of October and at the beginning of November, which corresponds to the beginning of the dry season in West Africa. The phenological appearance of C. africanus is in agreement with phenology of plant pathogenic fungi in other tropical regions (Piepenbring et al. 2015). The level of infection was very low, ranging from a single infected leaf to about 1 % of infected leaves on one particular tree. At the type locality in Ghana, two trees of B. costatum were growing side by side, at a distance of ca. 5 m, and only one of them was infected by C. africanus, though the level of infection was ca. 1 %, which was the highest observed infection among all detected locations of C. africanus. The locality near the Tanougou Waterfalls in Benin was visited in 2011 and 2012, and a weak infection (one leaf) of the small tree was observed only in 2011, while in the subsequent year, the same tree was healthy. This suggests that infection by C. africanus may not appear on the same tree/population every year.
Discussion
Ceraceosorus africanus differs somewhat from the generic characters outlined in the diagnosis of Ceraceosorus (Cunningham et al. 1976), notably in lacking internal thickenings of basidia and in having a distinct hymenium produced on an external subhymenial layer (that could be referred to as sterile external stroma). Cunningham et al. (1976) reported in the generic diagnosis that Ceraceosorus shows an indeterminate hymenial thickening, but it is difficult to conclude what they meant by this term. Moreover, Cunningham et al. (1976) differentiated Ceraceosorus from Dicellomyces as lacking a well-defined external stroma, but this character was not included in the generic diagnosis, only mentioned in the text. Considering the ontogenetical development and other aspects of morphology (intracellular hyphae, sorus-like fructifications at least in the initial stage of development, waxy and pinkish fructifications, morphology of basidia and basidiospores), as well as the same host genus (Bombax L.), both C. africanus and Ceraceosorus bombacis are however similar and should be considered con-generic. These divergent characters may therefore not be relevant at the generic level but useful to distinguish between C. africanus and C. bombacis. In addition, C. africanus is morphologically different from C. bombacis (Cunningham et al. 1976) in having somewhat longer and thinner basidia [48–82 × 3–5 μm vs. (20–)35–50(−85) × 3–6.5 μm], shorter sterigmata (up to 15 μm long vs. up to 22 μm long) and slightly longer and wider basidiospores [(6.5–)9–16(−18) × (3–)3.4–4.8(−5.2) μm vs. (5.5–)8.5–14(−18.5) × 2–4.5 μm)] and, most importantly, in having a different macroscopic symptoms of infection. The infection of leaves is apparently local in C. africanus, i.e., only single leaves on one tree are infected, but fructification covers large parts of the leaf surface and resembles leaf blight. In contrast, the infection of C. bombacis resembles leaf spot and comprises small light pink to brown spots scattered over the lower leaf surface (Bakshi et al. 1972; Fig. 5g–i). Although it is premature to generalize this observation based on two known species, this difference in macroscopic symptoms resembles those in Exobasidium Woronin (Begerow et al. 2002a; Piątek et al. 2012), where the same host plant may be infected by species producing leaf spots or leaf blights (as surculicolous or systemic infection), e.g., Exobasidium arescens Nannf. and Exobasidium myrtilli Siegm., respectively, on Vaccinium myrtillus L. (Ericaceae), and many others (Nannfeldt 1981).
The prominent and compact corticioid fructifications of Ceraceosorus africanus characterized by well-developed subhymenia and hymenia composed of dense palisades of basidioles and basidia, and resembling, for example, agaricomycotinous genera Gloeocystidiellum Donk, Phanerochaete P. Karst., or Phlebia Fr., represent a unique morphological trait in the subphylum Ustilaginomycotina. Indeed, some species of Exobasidium, like Exobasidium vaccinii (Fuckel) Woronin, form corticioid hymenia (Nannfeldt 1981), but not as prominent and well developed as in C. africanus. The fructification of other nonteliosporic Ustilaginomycotina species is rather small or loose and delicate. Obviously, this character cannot be considered an autapomorphy for C. africanus since this type of fructification evolved multiple times in different Basidiomycota lineages, most commonly in different clades of Agaricomycotina (corticioid fungi, Larsson 2007), but also in some species of Pucciniomycotina, e.g., in the rust Stereostratum corticioides (Berk. & Broome) H. Magn. (Cummins and Hiratsuka 2003), or in the atractiellomycete Saccoblastia farinacea (Höhn.) Donk (Bauer et al. 2006). However, it expands the structural diversity of sporulation within the Ustilaginomycotina.
The two currently known Ceraceosorus species are fully allopatric, separated both by geographic distribution and host plant species. Ceraceosorus africanus is known only on Bombax costatum in West Africa and Ceraceosorus bombacis occurs only on Bombax ceiba in India. The natural range of both host plants is widely separated and does not overlap in any area. B. costatum is naturally distributed in West Africa, while B. ceiba in tropical southern and eastern Asia, Indonesia, Philippines, Papua New Guinea, and northern Australia. Assuming that both Ceraceosorus species are strictly host species specific, it could be hypothesized that the speciation of C. africanus and C. bombacis was triggered by geographical isolation and ecological factors (host plant species).
The two currently known Ceraceosorus species are separated by a considerable genetic distance (uncorrected p values: 28S = 4.03 % or 83 bp, RPB2 = 18.5 % or 196 bp). Such a considerable genetic distance between Ceraceosorus africanus and Ceraceosorus bombacis could, e.g., be an indication for an ancient separation of the lineages under a model of neutral sequence evolution, or an indication for a selection of the loci under a nonneutral model of sequence evolution. Assuming an ancient separation of the lineages, the currently recognized species (1) may represent living descendants of a larger but extinct Ceraceosorus diversity, or (2) undiscovered species still may exist in poorly studied tropical countries, especially on the remaining Bombax species (Bombax buonopozense P. Beauv., Bombax insigne Wall., Bombax mossambicense A. Robyns; Malvaceae). The discovery of C. africanus, a common species producing conspicuous symptoms of infection on the common tree, Bombax costatum, but occurring in the phytopathologically unstudied Sudanian savanna biome of West Africa could support the latter assumption. Moreover, similar to other lineages of Ustilaginomycotina, saprobic yeast species may contribute to unknown Ceraceosorus diversity. In fact, one yeast species assigned to this genus was announced but not yet described (Kijpornyongpan and Aime 2014; Albu et al. 2015).
The discovery of Ceraceosorus africanus in West Africa raises the number of smut species parasitic on members of the Malvaceae. It also highlights the significance of scrutiny of the tropical ecosystems in order to increase the knowledge of fungal diversity and to find evolutionary unique “missing” fungal taxa in general (e.g., Isaac et al. 1993; Aime and Brearley 2012; Gazis et al. 2012) and smut fungi in particular. Previously, eight smut species were recognized on hosts of that family, including five teliosporic species from the genera Entyloma de Bary, Geminago Vánky & R. Bauer, and Pericladium Pass. (Table 3; Vánky 1996, 2011, 2012), and three nonteliosporic species from the genera Ceraceosorus and Volvocisporium Begerow, R. Bauer & Oberw. (Table 3; Cunningham et al. 1976; Begerow et al. 2001; Ritschel et al. 2008). All of these genera are known exclusively on hosts in the Malvaceae with the exception of Entyloma, which members infect hosts in multiple dicot families (Begerow et al. 2002b; Vánky 2012; Lutz and Piątek 2016), and only Entyloma sidae-rhombifoliae Cif. (Ciferri 1928, but doubtful smut species according to Zundel 1939) is parasitic on the Malvaceae. Interestingly, Ceraceosorus and Geminago are restricted to host species from different subfamilies, the Bombacoideae and the Helicteroideae, respectively, while Pericladium and Volvocisporium are both parasitic on members of the Grewioideae (Table 3). However, these smut genera are not phylogenetically related, indicating that in the evolution of smut fungi, hosts in Malvaceae were colonized several times from different ancestors. Interestingly, all smut species on hosts in the Malvaceae, except North American En. sidae-rhombifoliae (Vánky 2012), are confined to the African-Indian-Australian part of Gondwanaland, and Ceraceosorus and Volvocisporium have allopatric species in Africa and India. The disjunctive occurrence of smuts between Africa and India was already reported for some species (e.g., Piątek et al. 2014), but the mechanisms responsible for such a distribution pattern are not yet known.
The phylogenetic affinities of the Ceraceosorales, represented by Ceraceosorus bombacis, have previously been analyzed only few times (Begerow et al. 2006, the same analyses later repeated by Begerow et al. 2014; Albu et al. 2015; Sharma et al. 2015). In the multigene analyses, including ITS, 28S, ATP6, and β-tubulin sequences, conducted by Begerow et al. (2006), C. bombacis was nested within the Exobasidiomycetes occupying a common branch with Tilletiopsis albescens Gokhale and members of the Entylomatales, however without statistical support. In the multigene analyses, including 18S, ITS, 28S, TEF1, and β-tubulin sequences, conducted by Albu et al. (2015), the Ceraceosorales was weakly supported as member of the Ustilaginomycetes. Thus, the affinities of the Ceraceosorales were not resolved by these analyses (Begerow et al. 2006; Albu et al. 2015).
The molecular phylogenetic analyses of the single gene dataset (28S) conducted in this study (Fig. 3), including both Ceraceosorus africanus and Ceraceosorus bombacis, supported the monophyly of Ceraceosorus and confirmed the placement of this genus and the order Ceraceosorales within the Ustilaginomycotina in an unresolved relation to the Ustilaginomycetes and Exobasidiomycetes.
The molecular phylogenetic analyses of the multigene dataset (18S/5.8S/28S/RPB2/TEF1; Fig. 4) revealed Exobasidium rhododendri, i.e., member of the Exobasidiales, as the closest relative of Ceraceosorus, both clustering together with Entyloma calendulae, i.e., member of the Entylomatales. Thus, it is plausible that the Ceraceosorales belongs to the Exobasidiomycetes (sensu Begerow et al. 2006). It is noteworthy, however, that in this study, the Exobasidiomycetes were not resolved as monophyletic in both the single gene and the multigene analyses (Figs. 3 and 4). This is in congruence with several previous analyses made by, e.g., Begerow et al. (2006, dataset 18S/ITS/28S/ATP6/β-tubulin, their Supplementary Figure 1) or Wang et al. (2014). Interestingly, considering the ultrastructural characters studied by Bauer et al. (1997), the Exobasidiomycetes also have been divided into two groups: orders with species lacking an interaction apparatus (Georgefischeriales, Microstromatales, Tilletiales) and orders with species having an interaction apparatus (Entylomatales, Exobasidiales, Doassansiales), the latter referred to as superorder Exobasidianae. The ultrastructural characters of Ceraceosorus bombacis (Begerow et al. 2006, 2014), such as a local interaction zone with simple interaction apparatus, indicate its affinity to the superorder Exobasidianae. In this superorder, Exobasidiales and Doassansiales are characterized by having a complex interaction apparatus, while Entylomatales is characterized by having a simple interaction apparatus (Bauer et al. 1997; Begerow et al. 2006, 2014). Thus, the Ceraceosorales shares this ultrastructural trait only with the Entylomatales. This could suggest a close phylogenetic relation of the Ceraceosorales to the Entylomatales, which was also revealed (though statistically not supported) in the multigene analyses of Begerow et al. (2006, 2014), as well as inferred and well supported in the multigene analyses of the current study.
The superorder Exobasidianae has been resolved as monophyletic in the ITS/28S/ATP6/β-tubulin analysis of Begerow et al. (2006). In the current multigene analyses, the Exobasidianae formed a monophyletic group but without statistical support (Fig. 4). Thus, we are in favor of the following evolutionary scenario for the Ceraceosorales and the Exobasidiomycetes (sensu Begerow et al. 2006): (1) the order Ceraceosorales belongs to the superorder Exobasidianae and is related to the Entylomatales and the Exobasidiales; and (2) the class Exobasidiomycetes is composed of two major lineages, probably deserving their classification at the class level, defined by ultrastructural characters of cellular interactions. A robust phylogenetic hypothesis for this scenario may not be resolved by multigene phylogenetic analyses, reinforcing the similar conclusion of Begerow et al. (2006) for a different dataset. Probably, only phylogenomics may provide a robust hypothesis for the phylogeny of the Ustilaginomycotina. In recent phylogenomic analyses conducted by Sharma et al. (2015), including genomes of six species of the Ustilaginomycotina, all branches received maximum statistical support. Ceraceosorus bombacis was resolved as sister species to Ustilago maydis (DC.) Corda, Sporisorium reilianum (J.G. Kühn) Langdon & Full., Ustilago hordei (Pers.) Lagerh., and Melanopsichium pennsylvanicum Hirschh., while Malassezia globosa Midgley, E. Guého & J. Guillot was resolved as basal to all sampled species. The sampling by Sharma et al. (2015) included members of only three orders (Ustilaginales, Ceraceosorales, Malasseziales), and the inclusion of representatives of the remaining orders to phylogenomic analyses is a challenge for future studies.
References
Aime, M. C., & Brearley, F. Q. (2012). Tropical fungal diversity: closing the gap between species estimates and species discovery. Biodiversity and Conservation, 21, 2177–2180.
Albu, S., Toome, M., & Aime, M. C. (2015). Violaceomyces palustris gen. et sp. nov. and a new monotypic lineage, Violaceomycetales ord. nov. in Ustilaginomycetes. Mycologia, 107, 1193–1204.
Amend, A. (2014). From dandruff to deep-sea vents: Malassezia-like fungi are ecologically hyper-diverse. PLoS Pathogens, 10(8), e1004277.
Amin, R. M., Strobel, G. A., Vishnevetsky, M., Ren, Y., & Geary, B. (2010). Biologically active endophytic Quambalaria sp. from Leptospermum junipae in Australia. Mycology, 1, 67–74.
Bakkeren, G., Kronstad, J. W., & Levesque, C. A. (2000). Comparison of AFLP fingerprints and ITS sequences as phylogenetic markers in Ustilaginomycetes. Mycologia, 92, 510–521.
Bakshi, B.R., Reddy, M.A.R., Puri, Y.N., & Singh S. (1972[1973]). Forest Disease Survey Final Technical Report 1967–72. Dehra Dun: Publication of Forest Research Institute.
Bauer, R., Oberwinkler, F., & Vánky, K. (1997). Ultrastructural markers and systematics in smut fungi and allied taxa. Canadian Journal of Botany, 75, 1273–1314.
Bauer, R., Vánky, K., Begerow, D., & Oberwinkler, F. (1999). Ustilaginomycetes on Selaginella. Mycologia, 91, 475–484.
Bauer, R., Begerow, D., Vánky, K., & Oberwinkler, F. (2001). Georgefischeriales: a phylogenetic hypothesis. Mycological Research, 104, 416–424.
Bauer, R., Lutz, M., & Oberwinkler, F. (2005). Gjaerumia, a new genus in the Georgefischeriales (Ustilaginomycetes). Mycological Research, 109, 1250–1258.
Bauer, R., Begerow, D., Sampaio, J. P., Weiß, M., & Oberwinkler, F. (2006). The simple-septate basidiomycetes: a synopsis. Mycological Progress, 5, 41–66.
Bauer, R., Lutz, M., Piątek, M., Vánky, K., & Oberwinkler, F. (2007). Flamingomyces and Parvulago, new genera of marine smut fungi (Ustilaginomycotina). Mycological Research, 111, 1199–1206.
Bauer, R., Lutz, M., Begerow, D., Piątek, M., Vánky, K., Bacigálová, K., & Oberwinkler, F. (2008). Anther smut fungi on monocots. Mycological Research, 112, 1297–1306.
Bauer, R., Garnica, S., Oberwinkler, F., Riess, K., Weiß, M., & Begerow, D. (2015). Entorrhizomycota: a new fungal phylum reveals new perspectives on the evolution of Fungi. PLoS One, 10, e0128183.
Begerow, D., Bauer, R., & Oberwinkler, F. (1997[1998]). Phylogenetic studies on nuclear LSU rDNA sequences of smut fungi and related taxa. Canadian Journal of Botany, 75, 2045–2056.
Begerow, D., Bauer, R., & Boekhout, T. (2000). Phylogenetic placements of ustilaginomycetous anamorphs as deduced from nuclear LSU rDNA sequences. Mycological Research, 104, 53–60.
Begerow, D., Bauer, R., & Oberwinkler, F. (2001). Muribasidiospora: Microstromatales or Exobasidiales? Mycological Research, 105, 798–810.
Begerow, D., Bauer, R., & Oberwinkler, F. (2002a). The Exobasidiales: an evolutionary hypothesis. Mycological Progress, 1, 187–199.
Begerow, D., Lutz, M., & Oberwinkler, F. (2002b). Implications of molecular characters for the phylogeny of the genus Entyloma. Mycological Research, 106, 1392–1399.
Begerow, D., Stoll, M., & Bauer, R. (2006[2007]). A phylogenetic hypothesis of Ustilaginomycotina based on multiple gene analyses and morphological data. Mycologia, 98, 906–916.
Begerow, D., Schäfer, A. M., Kellner, R., Yurkov, A., Kemler, M., Oberwinkler, F., & Bauer, R. (2014). Ustilaginomycotina. In D. J. McLaughlin, & J. W. Spatafora (Eds), The Mycota. vol. 7. Part A. Systematics and evolution (2nd ed., pp. 295–329). Berlin: Springer.
Boekhout, T., Fell, J. W., & O’Donnell, K. (1995). Molecular systematics of some yeast-like anamorphs belonging to the Ustilaginales and Tilletiales. Studies in Mycology, 38, 175–183.
Boekhout, T., Theelen, B., Houbraken, J., Robert, V., Scorzetti, G., Gafni, A., Gerson, U., & Sztejnberg, A. (2003). Novel anamorphic mite-associated fungi belonging to the Ustilaginomycetes: Meira geulakonigii gen. nov., sp. nov., Meira argovae sp. nov. and Acaromyces ingoldii gen. nov., sp. nov. International Journal of Systematic and Evolutionary Microbiology, 53, 1655–1664.
Boekhout, T., Gildemacher, P., Theelen, B., Müller, W. H., Heijne, B., & Lutz, M. (2006). Extensive colonization of apples by smut anamorphs causes a new postharvest disorder. FEMS Yeast Research, 6, 63–76.
Brock, P. M., Döring, H., & Bidartondo, M. I. (2009). How to know unknown fungi: the role of a herbarium. New Phytologist, 181, 719–724.
Cabañes, F. J. (2014). Malassezia yeasts: how many species infect humans and animals? PLoS Pathogens, 10(2), e1003892.
Carris, L. M., Castlebury, L. A., Huang, G., Alderman, S. C., Luo, J., & Bao, X. (2007). Tilletia vankyi, a new species of reticulate-spored bunt fungus with non-conjugating basidiospores infecting species of Festuca and Lolium. Mycological Research, 111, 1386–1398.
Castlebury, L. A., Carris, L. M., & Vánky, K. (2005). Phylogenetic analysis of Tilletia and allied genera in order Tilletiales (Ustilaginomycetes; Exobasidiomycetidae) based on large subunit nuclear rDNA sequences. Mycologia, 97, 888–900.
Castresana, J. (2000). Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Molecular Biology and Evolution, 17, 540–552.
Chandra, A., & Huff, D. R. (2008). Salmacisia, a new genus of Tilletiales: reclassification of Tilletia buchloeana causing induced hermaphroditism in buffalograss. Mycologia, 100, 81–93.
Ciferri, R. (1928). Quarta contribuzione allo studio degli Ustilaginales. Annales Mycologici, 26, 1–68.
Clinton, G. P. (1906). Order Ustilaginales. North American Flora, 7, 1–82.
Cummins, G. B., & Hiratsuka, Y. (2003). Illustrated genera of rust fungi (3rd ed.). St Paul: American Phytopathological Society.
Cunningham, J. L., Bakshi, B. K., Lentz, P. L., & Gilliam, M. S. (1976). Two new genera of leaf-parasitic fungi (Basidiomycetidae: Brachybasidiaceae). Mycologia, 68, 640–654.
de Beer, Z. W., Begerow, D., Bauer, R., Pegg, G. S., Crous, P. W., & Wingfield, M. J. (2006). Phylogeny of the Quambalariaceae fam. nov., including important Eucalyptus pathogens in South Africa and Australia. Studies in Mycology, 55, 289–298.
de Wachter, R., Neefs, J. M., Goris, A., & Van de Peer, Y. (1992). The gene coding for small ribosomal subunit RNA in the basidiomycete Ustilago maydis contains a group I intron. Nucleic Acids Research, 20, 1251–1257.
Deadman, M. L., Al Sadi, A. M., Al Maqbali, Y. M., Farr, D. F., & Aime, M. C. (2011). Additions to the rust fungi (Pucciniales) from northern Oman. Sydowia, 63, 155–168.
Denchev, C. M., & Moore, R. T. (2009). Validation of Malasseziaceae and Ceraceosoraceae (Exobasidiomycetes). Mycotaxon, 110, 379–382.
Döring, H. (2003). Molekularsystematische Untersuchungen an Vertretern der pflanzenparasitischen Gattung Exobasidium (Basidiomycota). Thesis University of Bayreuth, Germany.
Döring, H., & Blanz, P. (2000). 18S rDNA-Analysen bei der Gattung Exobasidium. Hoppea, 61, 85–100.
Fell, J. W., Boekhout, T., Fonseca, Á., Scorzetti, G., & Statzell-Tallman, A. (2000). Biodiversity and systematics of basidiomycetous yeasts as determined by large-subunit rDNA D1/D2 domain sequence analysis. International Journal of Systematic and Evolutionary Microbiology, 50, 1351–1371.
Gargas, A., & Taylor, J. W. (1992). Polymerase chain reaction (PCR) primers for amplifying and sequencing 18S rDNA from lichenized fungi. Mycologia, 84, 589–592.
Gazis, R., Miądlikowska, J., Lutzoni, F., Arnold, A. E., & Chaverri, P. (2012). Culture-based study of endophytes associated with rubber trees in Peru reveals a new class of Pezizomycotina: Xylonomycetes. Molecular Phylogenetics and Evolution, 65, 294–304.
González, V., Vánky, K., Platas, G., & Lutz, M. (2007). Portalia gen. nov. (Ustilaginomycotina). Fungal Diversity, 27, 45–55.
Gorfer, M., Blumhoff, M., Klaubauf, S., Urban, A., Inselsbacher, E., Bandian, D., Mitter, B., Sessitsch, A., Wanek, W., & Strauss, J. (2011). Community profiling and gene expression of fungal assimilatory nitrate reductases in agricultural soil. The ISME Journal, 5, 1771–1783.
Hamamoto, M., Tamura, M., & Nakase, T. (2000). Emended descriptions of Tilletiopsis washingtonensis, Tilletiopsis cremea and Tilletiopsis lilacina. International Journal of Systematic and Evolutionary Microbiology, 50, 925–930.
Hendrichs, M., Begerow, D., Bauer, R., & Oberwinkler, F. (2005). The genus Anthracoidea (Basidiomycota, Ustilaginales): a molecular phylogenetic approach using LSU rDNA sequences. Mycological Research, 109, 31–40.
Hopple, J. S., & Vilgalys, R. (1999). Phylogenetic relationships in the mushroom genus Coprinus and dark-spored allies based on sequence data from the nuclear gene coding for the large ribosomal subunit RNA: divergent domains, outgroups, and monophyly. Molecular Phylogenetics and Evolution, 13, 1–19.
Isaac, S., Frankland, J. C., Watling, R., & Whalley, A. J. S. (Eds.). (1993). Aspects of tropical mycology. Cambridge: Cambridge University Press.
Katoh, K., & Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Molecular Biology and Evolution, 30, 772–780.
Katoh, K., Kuma, K., Toh, H., & Miyata, T. (2005). MAFFT version 5: improvement in accuracy of multiple sequence alignment. Nucleic Acids Research, 33, 511–518.
Kautmanová, I., Adamčik, S., Lizoň, P., & Jančovičová, S. (2012). Revision of taxonomic concept and systematic position of some Clavariaceae species. Mycologia, 104, 521–539.
Kijpornyongpan, T., & Aime, M. C. (2014). Three new species of yeasts belonging to Exobasidiomycetes. Inoculum, 65(3), 34.
Kirk, P. M., Cannon, P. F., David, J. C., & Stalpers, J. A. (2001). Dictionary of the fungi (9th ed.). Wallingford: CAB International.
Kolařík, M., Slávikova, E., & Pažoutova, S. (2006). The taxonomic and ecological characterisation of the clinically important heterobasidiomycete Fugomyces cyanescens and its association with bark beetles. Czech Mycology, 58, 81–98.
Kottke, I., Suárez, J. P., Herrera, P., Cruz, D., Bauer, R., Haug, I., & Garnica, S. (2010). Atractiellomycetes belonging to the ‘rust’ lineage (Pucciniomycotina) form mycorrhizae with terrestrial and epiphytic neotropical orchids. Proceedings of the Royal Society B: Biological Sciences, 277(1685), 1289–1298.
Larsson, K. H. (2007). Re-thinking the classification of corticioid fungi. Mycological Research, 111, 1040–1063.
Le Gac, M., Hood, M. E., Fournier, E., & Giraud, T. (2007). Phylogenetic evidence of host-specific cryptic species in the anther smut fungus. Evolution, 61, 15–26.
Lebel, T., & Castellano, M. A. (2002). Type studies of sequestrate Russulales II. Australian and New Zealand species related to Russula. Mycologia, 94, 327–354.
Liu, Y. J., & Hall, B. D. (2004). Body plan evolution of ascomycetes, as inferred from an RNA polymerase II phylogeny. Proceedings of the National Academy of Sciences of the United States of America, 101(13), 4507–4512.
Liu, Y. J., Whelen, S., & Hall, B. D. (1999). Phylogenetic relationships among ascomycetes: evidence from an RNA polymerase II subunit. Molecular Biology and Evolution, 16, 1799–1808.
Lutz, M., & Piątek, M. (2016). Phylogenetic placement, DNA barcoding, morphology and evidence for the spreading of Entyloma cosmi, a species attacking Cosmos bipinnatus in temperate climate gardens. European Journal of Plant Pathology. doi:10.1007/s10658-016-0874-1
Lutz, M., Piątek, M., Kemler, M., Chlebicki, A., & Oberwinkler, F. (2008). Anther smuts of Caryophyllaceae: molecular analyses reveal further new species. Mycological Research, 112, 1280–1296.
Lutz, M., Vánky, K., & Bauer, R. (2012a). Melanoxa, a new genus in the Urocystidales (Ustilaginomycotina). Mycological Progress, 11, 149–158.
Lutz, M., Vánky, K., & Piątek, M. (2012b). Shivasia gen. nov. for the Australasian smut Ustilago solida that historically shifted through five different genera. IMA Fungus, 3, 143–154.
Lutzoni, F., Kauff, F., Cox, C. J., McLaughlin, D., Celio, G., Dentinger, B., Padamsee, M., Hibbett, D., James, T. Y., Baloch, E., et al. (2004). Assembling the Fungal Tree of Life: progress, classification, and evolution of subcellular traits. American Journal of Botany, 91, 1446–1480.
Maddison, W.P., & Maddison, D.R. (2011). Mesquite: a modular system for evolutionary analysis. Version 2.75. Online available at http://mesquiteproject.org.
Maier, W., Khoza, T., Harmse, N., Wingfield, B. D., & Wingfield, M. J. (2006). A disease epidemic on Zizyphus mucronata in the Kruger National Park caused by Coniodictyum chevalieri. Studies in Mycology, 55, 279–288.
Matheny, P. B. (2005). Improving phylogenetic inference of mushrooms with RPB1 and RPB2 nucleotide sequences (Inocybe; Agaricales). Molecular Phylogenetics and Evolution, 35, 1–20.
Matheny, P. B., Gossmann, J. A., Zalar, P., Arun Kumar, T. K., & Hibbett, D. S. (2006). Resolving the phylogenetic position of the Wallemiomycetes: an enigmatic major lineage of Basidiomycota. Canadian Journal of Botany, 84, 1794–1805.
Matheny, P. B., Schoch, C. S., & Hibbett, D. S. (2007). Contributions of rpb2 and tef1 to the phylogeny of mushrooms and allies (Basidiomycota, Fungi). Molecular Phylogenetics and Evolution, 43, 430–451.
McTaggart, A. R., Shivas, R. G., Geering, A. D. W., Callaghan, B., Vánky, K., & Scharaschkin, T. (2012). Soral synapomorphies are significant for the systematics of the Ustilago-Sporisorium-Macalpinomyces complex (Ustilaginaceae). Persoonia, 29, 63–77.
Nannfeldt, J. A. (1981). Exobasidium, a taxonomic reassessment applied to the European species. Symbolae Botanicae Upsalienses, 23(2), 1–72.
Nasr, S., Soudi, M. R., Fazeli, S. A. S., Nguyen, H. D. T., Lutz, M., & Piątek, M. (2014). Expanding evolutionary diversity in the Ustilaginomycotina: Fereydouniaceae fam. nov. and Fereydounia gen. nov., the first urocystidalean yeast lineage. Mycological Progress, 13, 1217–1226.
Oberwinkler, F. (2012). Evolutionary trends in Basidiomycota. Stapfia, 96, 45–104.
Olive, L. S. (1945). A new Dacrymyces-like parasite of Arundinaria. Mycologia, 37, 543–552.
Olive, L. S. (1951). Taxonomic notes on Louisiana fungi. III. Additions to the Tremellales. Mycologia, 43, 677–690.
Paap, T., Burgess, T. I., McComb, J. A., Shearer, B. L., & Hardy, G. E. S. J. (2008). Quambalaria species, including Q. coyrecup sp. nov., implicated in canker and shoot blight diseases causing decline of Corymbia species in the southwest of Western Australia. Mycological Research, 112, 57–69.
Padhi, S., & Tayung, K. (2013). Antimicrobial activity and molecular characterization of an endophytic fungus, Quambalaria sp. isolated from Ipomoea carnea. Annals of Microbiology, 63, 793–800.
Paz, Z., Burdman, S., Gerson, U., & Sztejnberg, A. (2007). Antagonistic effects of the endophytic fungus Meira geulakonigii on the citrus rust mite Phyllocoptruta oleivora. Journal of Applied Microbiology, 103, 2570–2579.
Piątek, M., Lutz, M., & Welton, P. (2012). Exobasidium darwinii, a new Hawaiian species infecting endemic Vaccinium reticulatum in Haleakala National Park. Mycological Progress, 11, 361–371.
Piątek, M., Lutz, M., & Chater, A. O. (2013). Cryptic diversity in the Antherospora vaillantii complex on Muscari species. IMA Fungus, 4, 5–19.
Piątek, M., Piątek, J., & Yorou, N. S. (2014). Emended description and geographical distribution of Sporisorium elegantis (Ustilaginaceae), a species shared between West Africa and India. Phytotaxa, 175(3), 148–154.
Piątek, M., Lutz, M., & Yorou, N. S. (2015). A molecular phylogenetic framework for Anthracocystis (Ustilaginales), including five new combinations (inter alia for the asexual Pseudozyma flocculosa), and description of Anthracocystis grodzinskae sp. nov. Mycological Progress, 14, 88.
Piepenbring, M., Begerow, D., & Oberwinkler, F. (1999). Molecular sequence data assess the value of morphological characteristics for a phylogenetic classification of species of Cintractia. Mycologia, 91, 485–498.
Piepenbring, M., Stoll, M., & Oberwinkler, F. (2002). The generic position of Ustilago maydis, Ustilago scitaminea, and Ustilago esculenta (Ustilaginales). Mycological Progress, 1, 71–80.
Piepenbring, M., Espinosa, J., Saldaña, L., & Cáceres, O. (2010). New records, host plants, morphological and molecular data of Exobasidiales (Basidiomycota) from Panama. Nova Hedwigia, 91, 231–242.
Piepenbring, M., Hofmann, T. A., Miranda, E., Cáceres, O., & Unterseher, M. (2015). Leaf shedding and weather in tropical dry-seasonal forest shape the phenology of fungi—lessons from two years of monthly surveys in southwestern Panama. Fungal Ecology, 18, 83–92.
Ran, Y., He, X., Zhang, H., Dai, Y., Li, L., & Bulmer, G. S. (2008). Seborrheic dermatitis flare in a Dutch male due to commensal Malassezia furfur overgrowth. Medical Mycology, 46, 611–614.
Rehner, S. A., & Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-a sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia, 97, 84–98.
Riess, K., Oberwinkler, F., Bauer, R., & Garnica, S. (2013). High genetic diversity at the regional scale and possible speciation in Sebacina epigaea and S. incrustans. BMC Evolutionary Biology, 13, 102.
Riess, K., Bauer, R., Kellner, R., Kemler, M., Piątek, M., Vánky, K., & Begerow, D. (2015). Identification of a new order of root-colonising fungi in the Entorrhizomycota: Talbotiomycetales ord. nov. on eudicotyledons. IMA Fungus, 6, 129–133.
Ritschel, A., Begerow, D., & Oberwinkler, F. (2008). A new species of Volvocisporium from Namibia, V. grewiae sp. nov. (Microstromatales, Ustilaginomycota). Mycological Progress, 7, 1–5.
Ronquist, F., Teslenko, M., Van der Mark, P., Ayres, D. L., Darling, A., Höhna, S., Larget, B., Liu, L., Suchard, M. A., & Huelsenbeck, J. P. (2012). MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Systematic Biology, 61, 539–542.
Rosa, C. A., Jindamorakot, S., Limtong, S., Nakase, T., Lachance, M. A., Fidalgo-Jiménez, A., Daniel, H. M., Pagnocca, F. C., Inácio, J., & Morais, P. B. (2009). Synonymy of the yeast genera Moniliella and Trichosporonoides and proposal of Moniliella fonsecae sp. nov. and five new species combinations. International Journal of Systematic and Evolutionary Microbiology, 59, 425–429.
Rush, T. A., & Aime, M. C. (2013). The genus Meira: phylogenetic placement and description of a new species. Antonie van Leeuwenhoek, 103, 1097–1106.
Schillberg, S., Groß, P., & Tiburzy, R. (1995). Isolation and characterization of the EF-1 alpha gene of the filamentous fungus Puccinia graminis f. sp. tritici. Current Genetics, 27, 367–372.
Schoch, C. L., Seifert, K. A., Huhndorf, S., Robert, V., Spouge, J. L., Levesque, C. A., Chen, W., & Fungal Barcoding Consortium. (2012). Nuclear ribosomal internal transcribed spacer (ITS) region as a universal DNA barcode marker for Fungi. Proceedings of the National Academy of Sciences of the United States of America, 109, 6241–6246.
Schröter, J. (1889). Die Pilze Schlesiens. In F. Cohn (Ed.), Kryptogamen-Flora von Schlesien 3(1). Breslau: J.U. Kern’s Verlag.
Sharma, R., Xia, X. J., Riess, K., Bauer, R., & Thines, M. (2015). Comparative genomics including the early-diverging smut fungus Ceraceosorus bombacis reveals signatures of parallel evolution within plant and animal pathogens of fungi and oomycetes. Genome Biology and Evolution, 7, 2781–2798.
Sipiczki, M., & Kajdacsi, E. (2009). Jaminaea angkorensis gen. nov., sp. nov., a novel anamorphic fungus containing an S943 nuclear small-subunit rRNA group IB intron represents a basal branch of Microstromatales. International Journal of Systematic and Evolutionary Microbiology, 59, 914–920.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics, 30, 1312–1313.
Stoll, M., Begerow, D., & Oberwinkler, F. (2005). Molecular phylogeny of Ustilago, Sporisorium, and related taxa based on combined analyses of rDNA sequences. Mycological Research, 109, 342–356.
Swann, E. C., & Taylor, J. W. (1995). Phylogenetic perspectives on basidiomycete systematics: evidence from the 18S rRNA gene. Canadian Journal of Botany, 73, 862–868.
Takahashi, H., Sekiguchi, H., Ito, T., Sasahara, M., Hatanaka, N., Ohba, A., Hase, S., Ando, S., Hasegawa, H., & Takenaka, S. (2011). Microbial community profiles in intercellular fluid of rice. Journal of General Plant Pathology, 77, 121–131.
Takashima, M., & Nakase, T. (1996). A phylogenetic study of the genus Tilletiopsis, Tilletiaria anomala and related taxa based on the small subunit ribosomal DNA sequences. Journal of General and Applied Microbiology, 42, 421–429.
Tanaka, E., Shimizu, K., Imanishi, Y., Yasuda, F., & Tanaka, C. (2008). Isolation of basidiomycetous anamorphic yeast-like fungus Meira argovae found on Japanese bamboo. Mycoscience, 49, 329–333.
Vánky, K. (1996). Mycosyrinx and other pair-spored Ustilaginales. Mycoscience, 37, 173–185.
Vánky, K. (2001). The new classification of the smut fungi, exemplified by Australasian taxa. Australian Systematic Botany, 14, 385–394.
Vánky, K. (2011). The genus Pericladium (Ustilaginales). Pericladiaceae fam. nov. Mycologia Balcanica, 8, 147–152.
Vánky, K. (2012). Smut fungi of the world. St. Paul: APS Press.
Vánky, K., & Lutz, M. (2011). Tubisorus, a new genus of smut fungi (Ustilaginomycetes) for Sorosporium pachycarpum. Mycologia Balcanica, 8, 129–135.
Vánky, K., Lutz, M., & Shivas, R. G. (2006). Anomalomyces panici, new genus and species of Ustilaginomycetes from Australia. Mycologia Balcanica, 3, 119–126.
Vánky, K., Lutz, M., & Bauer, R. (2008). Floromyces, a new genus of Ustilaginomycotina. Mycotaxon, 104, 171–184.
Vánky, K., Shivas, R. G., Barrett, M. D., & Lutz, M. (2013). Eriocortex eriocauli, gen. et sp. nov. (Ustilaginomycetes) from Australia. Mycobiota, 1, 9–16.
Vilgalys, R., & Hester, M. (1990). Rapid genetic identification and mapping of enzymatically amplified ribosomal DNA from several Cryptococcus species. Journal of Bacteriology, 172, 4238–4246.
Wang, Q. M., Theelen, B., Groenewald, M., Bai, F. Y., & Boekhout, T. (2014). Moniliellomycetes and Malasseziomycetes, two new classes in Ustilaginomycotina. Persoonia, 33, 41–47.
White, T. J., Bruns, T. D., Lee, S., & Taylor, J. W. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In M. A. Innis, D. H. Gelfand, J. J. Sninsky, & T. J. White (Eds.), PCR protocols: a guide to methods and applications (pp. 315–322). San Diego: Academic.
Wingfield, B. D., Ericson, L., Szaro, T., & Burdon, J. J. (2004). Phylogenetic patterns in the Uredinales. Australasian Plant Pathology, 33, 327–335.
Zundel, G. L. (1939). Additions and corrections to Ustilaginales. North American Flora, 7(14), 971–1045.
Zundel, G.L. (1953). The Ustilaginales of the world. Pennsylvania State College School of Agriculture Department of Botany Contribution, 176, xi+1–410.
Acknowledgments
We thank Rebekka Ziegler for assistance in the molecular lab and Jolanta Piątek for drawings and preparing the map of Africa. We highly appreciate the Director of Forest Research Institute, Dehra Dun (India), for permission to reproduce the figure showing the leaf with infection by Ceraceosorus bombacis published by Bakshi et al. (1972). This research was financially supported by grants of the National Science Centre (NCN) of Poland (project no. N N303 414037 for the years 2009–2014), the German Research Foundation (DFG; project no BA 75/3-1), and the Volkswagen Foundation.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Piątek, M., Riess, K., Karasiński, D. et al. Integrative analysis of the West African Ceraceosorus africanus sp. nov. provides insights into the diversity, biogeography, and evolution of the enigmatic Ceraceosorales (Fungi: Ustilaginomycotina). Org Divers Evol 16, 743–760 (2016). https://doi.org/10.1007/s13127-016-0285-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-016-0285-3