Abstract
The Acoelomorpha is an animal group comprised by nearly 400 species of misleadingly inconspicuous flatworms. Despite this, acoelomorphs have been at the centre of a heated debate about the origin of bilaterian animals for 150 years. The animal tree of life has undergone major changes during the last decades, thanks largely to the advent of molecular data together with the development of more rigorous phylogenetic methods. There is now a relatively robust backbone of the animal tree of life. However, some crucial nodes remain contentious, especially the node defining the root of Bilateria. Some studies situate Acoelomorpha (and Xenoturbellida) as the sister group of all other bilaterians, while other analyses group them within the deuterostomes which instead suggests that the last common bilaterian ancestor directly gave rise to deuterostomes and protostomes. The resolution of this node will have a profound impact on our understanding of animal/bilaterian evolution. In particular, if acoelomorphs are the sister group to Bilateria, it will point to a simple nature for the first bilaterian. Alternatively, if acoelomorphs are deuterostomes, this will imply that they are the result of secondary simplification. Here, we review the state of this question and provide potential ways to solve this long-standing issue. Specifically, we argue for the benefits of (1) obtaining additional genomic data from acoelomorphs, in particular from taxa with slower evolutionary rates; (2) the development of new tools to analyse the data; and (3) the use of metagenomics or metatranscriptomics data. We believe the combination of these three approaches will provide a definitive answer as to the position of the acoelomorphs in the animal tree of life.
Similar content being viewed by others
Historically—and even today—the Turbellaria dominate much of our phylogenetic thinking on the lower Metazoa; hardly any other group of invertebrates has been accorded a position of comparable importance or been subjected to so many different interpretations
(Peter Ax, Relationships and phylogeny of the Turbellaria. In The Lower Metazoa. University of California Press, Berkeley, 1963, ed. E. C. Dougherty).
The phylum Acoelomorpha
Acoelomorpha is a group of bilaterally symmetric animals with an apparent morphological simplicity: they lack body cavities, corporal segmentation, circulatory and respiratory systems, nephridia or protonephridia and larval stages, and their digestive system only has one opening to the exterior. The Acoelomorpha have been included in the phylum Platyhelminthes since it was created (Gegenbaur 1859) and are divided in two major clades: acoels and nemertodermatids. Platyhelminthes were, at the same time, split in three major lineages: Acoelomorpha, Catenulida and Rhabditophora (Ehlers 1985). However, it was already pointed out that there were no clear, unequivocal synapomorphies to unite the three groups of Platyhelminthes (for a review, see Haszprunar 1996; Julian et al. 1986). The sister group relationship of Acoela and Nemertodermatida (that together form the clade Acoelomorpha) was based on somehow stronger arguments. These included the ciliary rootlet system, a peculiar duet spiral cleavage during the early stages of development, lack of nephridia, well-defined guts and through-gut, even though the lack of complex structures could be a plesiomorphic feature (Achatz et al. 2012). There are, as well, some important morphological differences between these two groups of worms, the acoels and the nemertodermatids. For example, acoel sperm has two flagella and their statocyst bear single lithocyte, while nemertodermatid sperm has a single flagellum and their statocysts hold two lithocytes. An in depth description of the morphology of acoelomorphs has been recently published by Achatz et al. (2012).
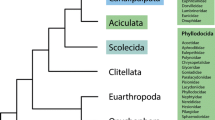
Despite the absence of clear synapomorphies to unite acoelomorphs and the rest of flatworms, most morphological phylogenetic studies positioned Acoelomorpha within Platyhelminthes, which appeared in different positions within Metazoa, usually within Protostomia (Fig. 1a, see references in Baguñà and Riutort 2004a). However, Haszprunar placed Acoelomorpha as the sister lineage to the rest of Bilateria (Haszprunar 1996) (Fig. 1b). In particular, Platyhelminthes was suggested to be a paraphyletic assemblage with Acoelomorpha as the earliest branching clade, followed by Rhabditophora, then Catenulida, and then the rest of Bilateria. This scenario is reminiscent of the planuloid-acoeloid hypothesis supported by Von Graff (Graff 1882) and Hyman (Hyman 1940), in which an acoel-like flatworm was suggested as the first bilaterian animal. In any case, the dominating hypotheses situated Acoelomorpha either within Platyhelminthes or as sister to Rhabditophora, Catenulida and the rest of Bilateria.
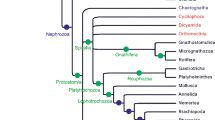
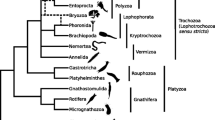
Diverse phylogenetic hypotheses on the Acoelomorpha through time. a Consensus tree of diverse morphological-based studies including acoelomorphs within Platyhelminthes in the protostomes (see text for references). b Tree modified from Haszprunar (1996) showing paraphyletic Platyhelminthes as early diverging bilaterians. c Summary of the first molecular phylogenies with a significative taxon sampling for Acoelomorpha and applying methodologies to overcome long-brach attraction (LBA) problems, based on diverse markers such as 18S rRNA, multiple genes, or phylogenomic approaches (see text for references). d Alternative phylogenomic hypothesis, placing acoelomorphs as sister to deuterostomes or within them, supported by Philippe et al. (2011a)
The possibility that Platyhelminthes, and specially Acoela and Nemertodermatida are an offshoot of the first bilaterians, is crucial to our view of animal evolution (Baguñà and Riutort 2004b). The planuloid-acoeloid proposal supports a simple last common ancestor of the bilaterians, similar to the planula larvae of cnidarians, that lacked coelom, segmentation, though-gut, larval stages and many organ systems. The competing hypothesis is the archicoelomate theory (Sedgwick 1884; Jagersten 1955), which suggests a rather complex last bilaterian ancestor with a coelom, through-gut and a complex nervous system. Thus, molecular data appeared as an ideal, independent dataset in which to test these contrasting hypotheses.
Molecular phylogenies situate Acoelomorpha as the sister-group to the rest of Bilateria and Platyhelminthes as lophotrochozoans
The first molecular phylogenies, based on the small subunit ribosomal gene or 18S (18S rRNA), rearranged the main lineages of the animal tree of life and pointed to a “new animal phylogeny” (Adoutte et al. 2000; Philippe and Telford 2006). A major change within the Bilateria was the definition of the superclades Ecdysozoa (Aguinaldo et al. 1997) and Lophotrochozoa (Halanych et al. 1995). Ecdysozoa included all animals whose body is encapsulated by an external cuticle that undergoes moulting (ecdysis) (Aguinaldo et al. 1997). Lophotrochozoa united animals with no clear morphological synapomorphies (as the name indicates, some sport a feeding structure called lophophore, others a trochophore larvae, others none of these) (Halanych et al. 1995). Other authors have used the term Spiralia to group some or all the members of the Lophotrochozoa, based on the idea that the last common ancestor or the group likely displayed spiral cleavage during development; however, this trait is not present in all the members of the clade and displays homoplasy as result of secondary simplification (for a review, see (Giribet 2008).
The first 18S rRNA molecular phylogeny including a flatworm situated Platyhelminthes as the earliest branching bilaterians (Field et al. 1988). This result was reproduced in more recent analyses (Winnepenninckk et al. 1995). This supported the classical morphological view placing Platyhelminthes as the sister group to the rest of Bilateria, and suggested an acoelomate-to-coelomate ladder-like evolution in bilaterians. The results, however, had to be taken with caution, since the 18S rRNA sequences of Platyhelminthes showed remarkably long-branches, indicating that they had higher rates of nucleotide substitution than other metazoans (Winnepenninckk et al. 1995). At that time, the “long-branch attraction artefact” (LBA) had been already described, in which taxa with longer branches tend to artifactually group together, usually incorrectly, because they cluster closer to the outgroup (Felsenstein 1978). LBA is a pervasive problem in molecular phylogenies, not just in the ribosomal rRNA genes but also in large phylogenomic datasets, where it often obscures the relationships between key taxa. Different approaches to overcome such systematic problems have been suggested in the literature (Anderson and Swofford 2004; Bergsten 2005; Paps et al. 2009a). Interestingly, in the first phylogenetic tree inferred with a wide taxon sampling of Platyhelminthes and other animal phyla, Platyhelminthes (without Acoela) appeared related to the Protostomia and not as the earliest-branching bilaterians (Carranza et al. 1997). However, the phylogenetic position of acoels was considered unreliable by the authors (Carranza et al. 1997), due to LBA. Indeed, the only two acoel sequences available at that time had even longer branches than those from other Platyhelminthes. Thus, although the first molecular data seemed to confirm that Platyhelminthes (likely including acoels) were the sister group to the rest of Bilateria, more robust, taxon-rich, analyses situated them within the protostomes, leaving the position of acoels unresolved due to their faster rates of nucleotide substitution.
The finding of acoels with slower rates of nucleotide substitution apparently settled the debate by overcoming the persistent LBA problem (Ruiz-Trillo et al. 1999). In a taxon-rich analysis, we screened up to 18 different acoel taxa and removed those with faster evolutionary rates. When only the sequence of an acoel with similar evolutionary rates than the rest of animals (Paratomella rubra) was used, acoels appeared consistently and with strong nodal support as the sister group to the other Bilateria (Ruiz-Trillo et al. 1999), far from the Platyhelminthes, which branched within Lophotrochozoa. Thus, a new evolutionary scenario emerged: acoels were situated as sister group to the rest of Bilateria, and Platyhelminthes as members of the Lophotrochozoa assemblage (Fig. 1c).
Confidence in the placement of acoels as a sister group to the Bilateria and away from the Platyhelminthes was boosted by subsequent studies that used ribosomal genes (Glenner et al. 2004; Jondelius et al. 2002; Mallatt et al. 2010; Telford et al. 2003; Wallberg et al. 2007) as well as further independent molecular markers, such as the myosin heavy chain (Ruiz-Trillo et al. 2002). Further confirmation came by creating concatenated multigene datasets, using either mitochondrial (Ruiz-Trillo et al. 2004; Mwinyi et al. 2010) or nuclear-coding genes (Paps et al. 2009b) (see references in Table 1). Those trees included nemertodematids as well, which appeared either branching after acoels as the sister group of the remaining bilaterians, or as the sister group to acoels, forming the Acoelomorpha clade. The position of acoelomorphs as early diverging bilaterians and their divorce from platyhelminths, has also been confirmed by molecular qualitative traits such as Hox genes (Cook et al. 2004; Jimenez-Guri et al. 2006), changes in the mitochondrial genetic code (Telford et al. 2000), miRNAs (Pasquinelli et al. 2003) and mtDNA gene rearrangements (Ruiz-Trillo et al. 2004). These compelling emerging data lead some authors to define Acoelomorpha and Platyhelminthes as two different metazoan phyla (Baguñà and Riutort 2004a).
The advent of large-scale sequencing (mainly by the use of expressed sequence taqs (ESTs)) allowed phylogenomic analyses of datasets composed of hundreds of concatenated genes. One of the first such analysis to include Acoela (Dunn et al. 2008) inferred acoels to be within Lophotrochozoa, as sister group to the Gnathostomulida, although with low nodal support (Table 1). The authors argued that the reason for the low statistical support on the phylogenetic position of acoels was their level of missing data. However, in a follow-up study that included substantially more data, acoelomorphs (acoels and nemertodermatids) were recovered as a sister group to the rest of Bilateria with relatively strong nodal support (Hejnol et al. 2009) (Table 1). Thus, different types of molecular data, from single gene and multigene phylogenies to qualitative characters, appeared to support the position of acoelomorphs as the sister group of the remaining Bilateria.
A contrasting hypothesis: acoelomorphs are deuterostomes
The position of acoelomorphs as sister group to the remaining bilaterians established by previous studies was challenged by two different studies. The first was a phylogenomic analysis in which new data for the acoel Convoluta pulchra was generated. The acoel representative appeared not as an early-branching bilaterian but as sister group to the deuterostomes (Philippe et al. 2007). The authors used a Bayesian approach combined with sophisticated models of evolution that should handle the challenges derived from LBA in large phylogenomic datasets. However, the position of the acoel lacked statistical support (see Table 1) and, thus, the authors concluded that its position remained unclear. Thus, this dataset corroborates that acoels were not Platyhelminthes, but weakly pointed to an alternative possibility in which acoels were related to deuterostomes, and additional data was needed.
It is worth mentioning here that there is an additional worm lineage that had been morphologically associated to acoelomorphs, the enigmatic Xenoturbella. This taxa was discovered by Sixten Bock and thoroughly described by Westblad in 1949 (Westblad 1949), who already suggested a platyhelminth nature for the worm (see also Lundin 1998). The first 18S rRNA from Xenoturbella turned out to be a contamination from a mollusc (Norén and Jondelius 1997), and given the absence of new material, Xenoturbella sequences were not included in the first studies dealing with the position of acoelomorphs. Subsequent uncontaminated ribosomal RNA analyses placed Xenoturbella surprisingly among the deuterostomes (Paps et al. 2009b; Philippe et al. 2007; Bourlat et al. 2003). However, later phylogenomic studies including EST data suggested Xenoturbella to be a sister group of Acoelomorpha together forming the sister group to the rest of bilaterians (Hejnol et al. 2009).
A definitive break from the line of studies suggesting acoelomorphs as the sister group to the rest of bilaterians came with a complete phylogenomic analyses that concluded that both Xenoturbella and Acoelomorpha (named the Xenacoelomorpha clade) were deuterostomes (Philippe et al. 2011a). This study considered four different lines of evidence: mitochondrial protein coding genes, nuclear-coding phylogenomic analyses (a subset of the genes used by Hejnol et al. 2009), microRNA content, and the absence/presence of the gene Rsb66. All these datasets provided weak evidence for the deuterostome affiliation of Acoelomorpha and Xenoturbella.
The nuclear-coding datasets merged data from Dunn et al. (2008) and Philippe et al. (2009), as well as Hejnol et al. (2009). The final alignments included a high number of ribosomal genes, which were discarded in previous analyses (Dunn et al. 2008); ribosomal datasets have been shown to be problematic to infer phylogenies (Nosenko et al. 2013). The nuclear-coding gene datasets were analysed using Bayesian inference, but instead of providing posterior probability values, the statistical support was assessed using a non-conventional approach: 100 bootstrap pseudo-replicates were produced and independently analysed with Phylobayes; then, the resulting trees were used to compute the bootstrap support values. The dynamics and comparative value of such method have not been broadly explored. Using this technique, the results show a 63 % of bootstrap support value for the deuterostome affiliation of Xenacoelomorpha (Philippe et al. 2011a). This is a relatively low support for a multigene analysis, which means that the position of Xenacoelomorpha among deutereostomes was indeed not statistically supported. Moreover, the single microRNA analysis run on this data (Dollo Parsimony) did not support the position of Acoelomorpha and Xenoturbella as deuterostomic lineages. Rather, the most parsimonious topology included Acoelomorpha and Xenoturbella as the sister group to the rest of Bilateria. Nonetheless, the authors proposed an ad hoc explanation, arguing that the inconsistent miRNA pattern could be explained by secondary miRNA gene loss related to the divergent nature of xenoacelomorphs. In any case, the use of microRNAs for phylogeny has recently been shown to be misleading due to complex evolutionary histories of miRNA families, including prevalent secondary losses and very different evolutionary rates depending on the group analysed (Thomson et al. 2014).
Another line of evidence was the presence of the gene Rsb66, which was shown to be shared exclusively by deuterostomes, Acoelomorpha and Xenoturbella, and thus considered a molecular synapomorphy (see also the epimerase gene (de Mendoza and Ruiz-Trillo 2011)). However, any traits linking the Acoelomorpha with Deuterostomes can be also interpreted as an ancestral bilaterian feature, secondarily lost in the protostome lineage. Other such putative cases of unique genome signatures have later on shown to be cases of secondary losses, once a wider taxon sampling is analysed (Shadwick and Ruiz-Trillo 2012).
The authors were aware of all these problems with their datasets, but emphasised the consistent patterns emerging from these four independent datasets. In our view, the support for each dataset in the analysis of Philippe et al. was so weak that the potential consistent pattern remains weak as well.
A recent study focused on a ctenophore genome and the phylogenetic position of comb jellies also included Acoelomorpha and Xenoturbella in its EST-based datasets (Ryan et al. 2013). The authors performed thorough analyses with both maximum likelihood and Bayesian inference approaches combined with sophisticated models of evolution, and different taxon sampling as outgroups. Most of the ML trees in that paper showed a monophyletic Acoelomorpha as the sister group to the rest of Bilateria. The Bayesian analyses inferred paraphyletic deuterostomes as early diverging bilaterians and acoelomorphs as sister to protostomes (Table 1). It is worth mentioning that the Bayesian trees in this study (performed with the CAT model) did not converge due to the large size of the dataset.
Finally, Srivastava et al. (2014) published a study on the regeneration mechanisms of acoels which included extensive phylogenomic analyses focused on resolving the position of Acoelomorpha and Xenoturbella. All the ML and Bayerian analyses, which used GTR or WAG models, displayed Acoelomorpha as the sister group to the rest of Bilateria, while the Bayesian tress generated using CAT or CAT + GTR models recovered paraphyletic deuterostomes and acoelomorphs as sister to protostomes (Table 1). Importantly, none of the analyses of Ryan et al. (2013) or Srivastava et al. (2014) showed Acoelomorpha as sister group or within Deuterostomia.
The fact that large phylogenomics datasets have not confidently resolved this issue raises some concerns. It is clear that the explosion of phylogenomic datasets has greatly revitalised the study of evolutionary relationships, but it has also spawned a discussion as to the relative value of the new data and the new methods. The addition of sufficiently large amounts of information—both new markers and new taxa—would seem intuitively to be sufficient to clarify previously problematic phylogenetic relationships. However, some authors highlight the importance, as well, of better methods of analysis and of the paramount importance of data quality (Nosenko et al. 2013; Osigus et al. 2013; Philippe et al. 2011b). Some studies have revealed that certain problems that lead to systematic errors can be exacerbated by high-throughput approaches, including contaminations, missing data, LBA, paralogy concerns, etc. (Roure et al. 2013). Probably, the solution lies between the two camps: while having as much data as possible is important, sifting the quality of this data and analysing it with appropriate methods is also needed.
Are acoelomorphs simple or simplified?
Molecular phylogenies have so far provided two alternative scenarios for the position of Acoelomorpha (with or without Xenoturbella, Table 1): either they are the sister group to the rest of Bilateria (sometimes named Eubilateria (Ax 2012) or Nephrozoa (Jondelius et al. 2002)) or they are deuterostomes. The two models have very different implications for the evolution of animals. Thus, what are the consequences of one versus the other? Are their body plans ancestrally simple (originally lacking traits that are present in the other bilaterians), or secondarily simplified (secondarily loosing characters that were present in the last common ancestor of all bilaterians)? There are three possible scenarios derived from these two phylogenetic positions.
The placement of Acoelomorpha as the sister group to the rest of Bilateria seems, at first, to lend support to a simple first bilaterian in line with the planuloid-acoeloid theory. While extant acoelomorphs would have evolved independently from other bilaterians for hundreds of millions of years, many of their traits could be considered a good proxy for the character states of the most recent common bilaterian ancestor: simple morphology, the lack of excretory and vascular system, the simple brain, direct development, and blind gut.
In contrast, the phylogenies pointing to a phylogenetic affinity with deuterostomes are more difficult to interpret, as some point to a sister group relationship with all the deuterostomes while others place acoelomorphs within the deuterostomes (Table 1, footnotes). In the first case, there is the possibility that acoelomorphs have never possessed any of the features that strongly define deuterostomes from a morphological point of view (Turbeville et al. 1994). While phylogenetically acoelomorphs would not be the sister group to the rest of bilaterians, their grade of organisation could still be reminiscent of the last bilaterian common ancestor and their morphology could still be ancestrally simple. In contrast, their position as a sister group to echinoderms and hemichordates prompts to consider them as simplified deuterostomes. They would have lost many of the complex traits currently argued to be present in the last common ancestor of deuterostomes: coelomes, body compartimentalisation, complex organ systems (nervous, circulatory, respiratory, and excretory), digestive system with mouth and anus, gill slits, etc.
There is a third intriguing possibility! The position of acoelomorphs within deuterostomes renders them as simplified deuterostomes, but the same cannot be said about their position as early diverging bilaterians. As indicated above, this placement intuitively points to a simple condition (McShea 1991); however, a secondarily simplified body plan, even if less parsimonious, cannot be ruled out. No matter the nature of the first bilaterian, simple or complex, acoelomorphs could have followed a route of simplified evolution during all those hundreds of millions of years apart from their bilaterians brothers and sisters, losing traits that may have been present in the last common ancestor of the Bilateria (i.e. coelom, through-gut, etc.). To sum up, acoelomorphs as first splitting bilaterians or sister group to deuterostomes could be ancestrally simple or secondarily simplified, while their position within deuterostomes grants their simplified status.
The question is whether acoels are ancestrally simple bilaterians or secondarily simplified from a more complex bilaterian ancestor. This is difficult to answer given that it is epistemologically impossible to prove the absence of a structure that never was there. On the other hand, anatomical simplification is usually accompanied by vestigial traces of the trait lost. While progenesis has been often invoked before to justify some body simplifications (Rieger 1985), morphological or molecular traces of complex ancestry are found in the morphology or genomes of simplified organisms or structures (a good example are the diversity of mitochondrion related organelles (Clark and Roger 1995; Roger and Silberman 2002)). No such traces have been reported so far for acoelomorphs and deuterostomes, which is striking given the strong morphological signal in the set of characters defining deuterostomes. A recently reported enteropneust miniaturised to a size similar of many acoelomorphs (around 500 μm), claimed to be the product of progenetic processes, still displays iconic deuterostome traits such as gill slits or coelom (Worsaae et al. 2012). Complete genomes of acoelomorphs, which are presently lacking, could shed new light, maybe by finding informative rare genomic changes (Rokas and Holland 2000) and other molecular signatures. However, the matter is not simple. For example, and as pointed above, features shared between acoelomorphs and deuterostomes could be interpreted as bilaterian plesiomorphies lost in the protostome lineage. We believe, however, that careful comparative genomic analyses could be able to discriminate between a simple and simplified state of acoelomorphs.
Perspectives
To unravel whether Acoelomorpha (and perhaps Xenoturbella) occupy a pivotal position between non-bilaterian and bilaterian animals or have a deuterostome affiliation is a key to understanding the evolution of animals. The question is whether there is a way to settle this dispute.
Morphology, molecular phylogenies and rare genomic changes do not seem to converge on a single answer. A potential avenue of research could be a blind, systematic approach to find all genome-wide rare genomic changes throughout the animal tree with the widest possible taxonomic sampling. This may provide an unbiased view of what this kind of data supports. There is also a persistent problem with the long branches of acoels. The acoel sequences appear with extremely long branches in the phylogenomics trees. In this regard, it is worth pointing that the branch on the acoel P. rubra in both our 18S rRNA and 13-gene animal phylogeny (Paps et al. 2009b; Ruiz-Trillo et al. 1999) appeared shorter than other animal clades. The lack of P. rubra in phylogenomic analyses conducted thus far is discouraging and we believe efforts to obtain additional molecular data from P. rubra, or other acoel taxa with slower evolutionary rates, should be encouraged. We believe the time has come for the community to embrace additional technologies, such as using metagenomics or metatranscriptomics (i.e., the use of acoel sequences coming from the environment), to look for additional and independent data. Those novel approaches have proven to be very powerful to attack other major transitions in the tree of life (Rinke et al. 2013; Spang et al. 2015).
Conclusions
Molecular data has indeed significantly increased the resolution of the tree of animals. However, the position therein of the acoelomorphs remains an enigma, and one with too important evolutionary implications to leave it this way. The support for acoelomorphs as sister group to the remaining bilaterians is strong, but we cannot discard conflicting evidence (even if currently relatively weak) for their deuterostome affiliation. We thus that believe efforts should be directed towards developing new tools and to generate additional molecular data from acoelomorphs.
References
Achatz, J. G., Chiodin, M., Salvenmoser, W., Tyler, S., & Martinez, P. (2012). The Acoela: on their kind and kinships, especially with nemertodermatids and xenoturbellids (Bilateria incertae sedis). Organisms Diversity & Evolution, 13(2), 267–286. doi:10.1007/s13127-012-0112-4.
Adoutte, A., Balavoine, G., Lartillot, N., Lespinet, O., Prud’homme, B., & de Rosa, R. (2000). The new animal phylogeny: reliability and implications. Proceedings of the National Academy of Sciences of the United States of America, 97(9), 4453–4456.
Aguinaldo, A. M., Turbeville, J. M., Linford, L. S., Rivera, M. C., Garey, J. R., Raff, R. A., & Lake, J. A. (1997). Evidence for a clade of nematodes, arthropods and other moulting animals. Nature, 387(6632), 489–493. doi:10.1038/387489a0.
Anderson, F. E., & Swofford, D. L. (2004). Should we be worried about long-branch attraction in real data sets? Investigations using metazoan 18S rDNA. Molecular Phylogenetics and Evolution, 33(2), 440–451. doi:10.1016/j.ympev.2004.06.015.
Ax, P. (2012). Multicellular animals. Berlin: Springer Science & Business Media. doi:10.1007/978-3-642-80114-3.
Baguñà, J., & Riutort, M. (2004a). Molecular phylogeny of the Platyhelminthes. Canadian Journal of Zoology, 82(2), 168–193. doi:10.1139/z03-214.
Baguñà, J., & Riutort, M. (2004b). The dawn of bilaterian animals: the case of acoelomorph flatworms. Bioessays, 26(10), 1046–1057. doi:10.1002/bies.20113.
Bergsten, J. (2005). A review of long-branch attraction. Cladistics, 21(2), 163–193.
Bourlat, S. J., Nielsen, C., Lockyer, A. E., Littlewood, D. T. J., & Telford, M. J. (2003). Xenoturbella is a deuterostome that eats molluscs. Nature, 424(6951), 925–928. doi:10.1038/nature01851.
Carranza, S., Baguna, J., & Riutort, M. (1997). Are the Platyhelminthes a monophyletic primitive group? An assessment using 18S rDNA sequences. Molecular and Biological Evolution, 14(5), 485–497.
Clark, C. G., & Roger, A. J. (1995). Direct evidence for secondary loss of mitochondria in Entamoeba histolytica. Proceedings of the National Academy of Sciences of the United States of America, 92(14), 6518–6521.
Cook, C. E., Jiménez, E., Akam, M., & Saló, E. (2004). The Hox gene complement of acoel flatworms, a basal bilaterian clade. Evolution and Development, 6(3), 154–163. doi:10.1111/j.1525-142X.2004.04020.x.
de Mendoza, A., & Ruiz-Trillo, I. (2011). The mysterious evolutionary origin for the GNE gene and the root of bilateria. Molecular and Biological Evolution, 28(11), 2987–2991. doi:10.1093/molbev/msr142.
Dunn, C. W., Hejnol, A., Matus, D. Q., Pang, K., Browne, W. E., Smith, S. A., et al. (2008). Broad phylogenomic sampling improves resolution of the animal tree of life. Nature, 452(7188), 745–749. doi:10.1038/nature06614.
Ehlers, U. (1985). Das phylogenetische System der Plathelminthes. Stuttgart.
Felsenstein, J. (1978). Cases in which parsimony or compatibility methods will be positively misleading. Systematic Zoology, 27, 401–410.
Field, K. G., Olsen, G. J., Lane, D. J., Giovannoni, S. J., Ghiselin, M. T., Raff, E. C., et al. (1988). Molecular phylogeny of the animal kingdom. Science, 239(4841 Pt 1), 748–753.
Gegenbaur, C. (1859). Grundzüge der Vergleichenden Anatomie. (W. Engelmann, Ed.). Leipzig.
Giribet, G. (2008). Assembling the lophotrochozoan (=spiralian) tree of life. Philosophical Transactions of the Royal Society, 363(1496), 1513–1522.
Glenner, H., Hansen, A. J., Sørensen, M. V., Ronquist, F., Huelsenbeck, J. P., & Willerslev, E. (2004). Bayesian inference of the metazoan phylogeny. Current Biology, 14(18), 1644–1649. doi:10.1016/j.cub.2004.09.027.
Graff, L. V. (1882). Monographie der Turbellarien. Leipzig: I. Rhabdocoelida.
Halanych, K. M., Bacheller, J. D., Aguinaldo, A. M., Liva, S. M., Hillis, D. M., & Lake, J. A. (1995). Evidence from 18S ribosomal DNA that the lophophorates are protostome animals. Science, 267(5204), 1641–1643.
Haszprunar, G. (1996). Plathelminthes and plathelminthomorpha-paraphyletic taxa. Journal of Zoological Systematics and Evolutionary Research, 34, 41–48.
Hejnol, A., Obst, M., Stamatakis, A., Ott, M., Rouse, G. W., Edgecombe, G. D., et al. (2009). Assessing the root of bilaterian animals with scalable phylogenomic methods. Proceedings of the Biological Sciences, 276(1677), 4261–4270. doi:10.1098/rspb.2009.0896.
Hyman, L. H. (1940). The Invertebrates: Protozoa Through Ctenophora: Vol-1 (Vol. 1). New York.
Jagersten, G. (1955). On the early phylogeny of the Metazoa: the bilaterogastraea theory. Zoologiska Bidrag Uppsala, 30, 321–354.
Jimenez-Guri, E., Paps, J., Garcia-Fernandez, J., & Saló, E. (2006). Hox and ParaHox genes in Nemertodermatida, a basal bilaterian clade. The International Journal of Developmental Biology, 50(8), 675–679. doi:10.1387/ijdb.062167ej.
Jondelius, U., Ruiz-Trillo, I., Bagu, A. J., & Riutort, M. (2002). The Nemertodermatida are basal bilaterians and not members of the Platyhelminthes. Zoologica Scripta, 31(2), 201–215.
Julian, P. S., Smith, I., Teyler, S., & Rieger, R. M. (1986). Is the Turbellaria polyphyletic? Hydrobiologia, 132, 13–21.
Lundin, K. (1998). The epidermal ciliary rootlets of Xenoturbella bocki (Xenoturbellida) revisited: new support for a possible kinship with the Acoelomorpha (Platyhelminthes). Zoologica Scripta, 27(3), 263–270. doi:10.1111/j.1463-6409.1998.tb00440.x.
Mallatt, J., Craig, C. W., & Yoder, M. J. (2010). Nearly complete rRNA genes assembled from across the metazoan animals: effects of more taxa, a structure-based alignment, and paired-sites evolutionary models on phylogeny reconstruction. Molecular Phylogenetics and Evolution, 55(1), 1–17. doi:10.1016/j.ympev.2009.09.028.
McShea, D. W. (1991). Complexity and evolution: what everybody knows. Biology and Philosophy, 6, 303–324.
Mwinyi, A., Bailly, X., Bourlat, S. J., Jondelius, U., Littlewood, D. T., & Podsiadlowski, L. (2010). The phylogenetic position of Acoela as revealed by the complete mitochondrial genome of Symsagittifera roscoffensis. BMC Evolutionary Biology, 10, 309. doi:10.1186/1471-2148-10-309.
Norén, M., & Jondelius, U. (1997). Xenoturbella ’s molluscan relativest. Nature, 390(6655), 31–32. doi:10.1038/36242.
Nosenko, T., Schreiber, F., Adamska, M., Adamski, M., Eitel, M., Hammel, J., et al. (2013). Deep metazoan phylogeny: when different genes tell different stories. Molecular Phylogenetics and Evolution, 67(1), 223–233. doi:10.1016/j.ympev.2013.01.010.
Osigus, H.-J., Eitel, M., & Schierwater, B. (2013). Chasing the urmetazoon: striking a blow for quality data? Molecular Phylogenetics and Evolution, 66(2), 551–557. doi:10.1016/j.ympev.2012.05.028.
Paps, J., Baguñà, J., & Riutort, M. (2009a). Lophotrochozoa internal phylogeny: new insights from an up-to-date analysis of nuclear ribosomal genes. Proceedings of the Biological Sciences, 276(1660), 1245–1254. doi:10.1098/rspb.2008.1574.
Paps, J., Baguñà, J., & Riutort, M. (2009b). Bilaterian phylogeny: a broad sampling of 13 nuclear genes provides a new Lophotrochozoa phylogeny and supports a paraphyletic basal acoelomorpha. Molecular and Biological Evolution, 26(10), 2397–2406. doi:10.1093/molbev/msp150.
Pasquinelli, A. E., McCoy, A., Jiménez, E., Saló, E., Ruvkun, G., Martindale, M. Q., & Baguñà, J. (2003). Expression of the 22 nucleotide let-7 heterochronic RNA throughout the Metazoa: a role in life history evolution? Evolution and Development, 5(4), 372–378.
Philippe, H., & Telford, M. J. (2006). Large-scale sequencing and the new animal phylogeny. Trends in Ecology & Evolution, 21(11), 614–620.
Philippe, H., Brinkmann, H., Martinez, P., Riutort, M., & Baguna, J. (2007). Acoel flatworms are not platyhelminthes: evidence from phylogenomics. PLoS ONE, 2(8), e717. doi:10.1371/journal.pone.0000717.
Philippe, H., Derelle, R., Lopez, P., Pick, K., Borchiellini, C., Boury-Esnault, N., et al. (2009). Phylogenomics revives traditional views on deep animal relationships. Current Biology, 19(8), 706–712.
Philippe, H., Brinkmann, H., Copley, R. R., Moroz, L. L., Nakano, H., Poustka, A. J., et al. (2011a). Acoelomorph flatworms are deuterostomes related to Xenoturbella. Nature, 470(7333), 255–258. doi:10.1038/nature09676.
Philippe, H., Brinkmann, H., Lavrov, D. V., Littlewood, D. T., Manuel, M., Worheide, G., & Baurain, D. (2011b). Resolving difficult phylogenetic questions: why more sequences are not enough. PLoS Biology, 9(3), e1000602. doi:10.1371/journal.pbio.1000602.
Rieger, R. (1985). The phylogenetic status of the acoelomate organisation within the Bilateria: A histological prespective. In S. C. Morris, R. Gibson, & H. M. Platt (Eds.), The Origin and relationship of lower invertebrates (pp. 101–122). Oxford.
Rinke, C., Schwientek, P., Sczyrba, A., Ivanova, N. N., Anderson, I. J., Cheng, J. F., et al. (2013). Insights into the phylogeny and coding potential of microbial dark matter. Nature, 499(7459), 431–437. doi:10.1038/nature12352.
Roger, A. J., & Silberman, J. D. (2002). Cell evolution: mitochondria in hiding. Nature, 418(6900), 827–829.
Rokas, A., & Holland, P. (2000). Rare genomic changes as a tool for phylogenetics. Trends in Ecology & Evolution, 15(11), 454–459.
Roure, B., Baurain, D., & Philippe, H. (2013). Impact of missing data on phylogenies inferred from empirical phylogenomic data sets. Molecular and Biological Evolution, 30(1), 197–214. doi:10.1093/molbev/mss208.
Ruiz-Trillo, I., Riutort, M., Littlewood, D. T., Herniou, E. A., & Baguna, J. (1999). Acoel flatworms: earliest extant bilaterian Metazoans, not members of Platyhelminthes. Science, 283(5409), 1919–1923.
Ruiz-Trillo, I., Paps, J., Loukota, M., Ribera, C., Jondelius, U., Baguna, J., & Riutort, M. (2002). A phylogenetic analysis of myosin heavy chain type II sequences corroborates that Acoela and Nemertodermatida are basal bilaterians. Proceedings of the National Academy of Sciences of the United States of America, 99(17), 11246–11251.
Ruiz-Trillo, I., Riutort, M., Fourcade, H. M., Baguna, J., & Boore, J. L. (2004). Mitochondrial genome data support the basal position of Acoelomorpha and the polyphyly of the Platyhelminthes. Molecular Phylogenetics and Evolution, 33(2), 321–332.
Ryan, J. F., Pang, K., Schnitzler, C. E., Nguyen, A. D., Moreland, R. T., Simmons, D. K., et al. (2013). The genome of the ctenophore Mnemiopsis leidyi and its implications for cell type evolution. Science, 342(6164), 1242592. doi:10.1126/science.1242592.
Sedgwick, A. (1884). Memoirs: on the origin metameric segmentation and some other morphological question. The Quarterly Journal of Microscopical Science, 24, 43–82.
Shadwick, J. D., & Ruiz-Trillo, I. (2012). A genomic survey shows that the haloarchaeal type tyrosyl tRNA synthetase is not a synapomorphy of opisthokonts. European Journal of Protistology, 48(1), 89–93. doi:10.1016/j.ejop.2011.10.003.
Spang, A., Saw, J. H., Jørgensen, S. L., Zaremba-Niedzwiedzka, K., Martijn, J., Lind, A. E., et al. (2015). Complex archaea that bridge the gap between prokaryotes and eukaryotes. Nature. doi:10.1038/nature14447.
Srivastava, M., Mazza-Curll, K. L., van Wolfswinkel, J. C., & Reddien, P. W. (2014). Whole-body acoel regeneration is controlled by Wnt and Bmp-Admp signaling. Current Biology, 24(10), 1107–1113. doi:10.1016/j.cub.2014.03.042.
Telford, M. J., Herniou, E. A., Russell, R. B., & Littlewood, D. T. (2000). Changes in mitochondrial genetic codes as phylogenetic characters: two examples from the flatworms. Proceedings of the National Academy of Sciences of the United States of America, 97(21), 11359–11364. doi:10.1073/pnas.97.21.11359.
Telford, M. J., Lockyer, A. E., Cartwright-Finch, C., & Littlewood, D. T. J. (2003). Combined large and small subunit ribosomal RNA phylogenies support a basal position of the acoelomorph flatworms. Proceedings of the Biological Sciences, 270(1519), 1077–1083. doi:10.1098/rspb.2003.2342.
Thomson, R. C., Plachetzki, D. C., Mahler, D. L., & Moore, B. R. (2014). A critical appraisal of the use of microRNA data in phylogenetics. Proceedings of the National Academy of Sciences of the United States of America, 111(35), E3659–E3668. doi:10.1073/pnas.1407207111.
Turbeville, J. M., Schulz, J. R., & Raff, R. A. (1994). Deuterostome phylogeny and the sister group of the chordates: evidence from molecules and morphology. Molecular and Biological Evolution, 11(4), 648–655.
Wallberg, A., Curini-Galletti, M., Ahmadzadeh, A., & Jondelius, U. (2007). Dismissal of acoelomorpha: acoela and nemertodermatida are separate early bilaterian clades. Zoologica Scripta, 36(5), 509–523.
Westblad, E. (1949). Xenoturbella bocki ng, n. sp., a peculiar, primitive turbellarian type (Vol. 1, pp. 3–29). Arkiv för zoologi.
Winnepenninckk, B., Backeljau, T., & De Wachter, R. (1995). Phylogeny of protostome worms derived from 18S rRNA sequences. Molecular and Biological Evolution, 12(4), 641–649.
Worsaae, K., Sterrer, W., Kaul-Strehlow, S., Hay-Schmidt, A., & Giribet, G. (2012). An anatomical description of a miniaturized acorn worm (hemichordata, enteropneusta) with asexual reproduction by paratomy. PLoS ONE, 7(11), e48529. doi:10.1371/journal.pone.0048529.
Acknowledgments
This work was supported by an Institució Catalana de Recerca i Estudis Avançats contract and a grant (BFU2014-57779-P) from Ministerio de Economia y Competitividad (MINECO) to IR-T. IR-T also acknowledges financial support from the Secretaria d’Universitats i Recerca del Departament d’Economia i Coneixement de la Generalitat de Catalunya (Project 2014 SGR 619). JP acknowledges support from the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013)/ERC grant [268513]. We thank Marta Riutort and Jaume Baguñà for comments to the manuscript.
Author information
Authors and Affiliations
Corresponding author
Additional information
This article is part of the Special Issue The new animal phylogeny: The first 20 years
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Ruiz-Trillo, I., Paps, J. Acoelomorpha: earliest branching bilaterians or deuterostomes?. Org Divers Evol 16, 391–399 (2016). https://doi.org/10.1007/s13127-015-0239-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13127-015-0239-1