Abstract
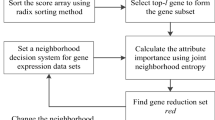
The classification of gene expression data provides a basis for the study of pathogenesis and treatment. However, this type of data is characterized by high dimensionality and small samples, which seriously affect the classification results. Consequently, it is necessary to use a gene selection algorithm to select key genes from gene expression data to improve the classification results, but the existing gene selection algorithm has the problems of low classification precision and high time complexity. Therefore, this paper proposes a gene selection algorithm using neighborhood uncertainty measures and Fisher score. First, to make full use of the information provided by the neighborhood decision system, the neighborhood fusion coverage and neighborhood fusion credibility are defined based on the neighborhood coverage and neighborhood credibility, and they are used to characterize neighborhood uncertainty measures. Second, the neighborhood uncertainty measures are extended by combining the algebraic and information theory views, and a heuristic nonmonotonic gene selection algorithm is designed based on the neighborhood uncertainty measures. The algorithm makes full use of the information in the neighborhood decision system to evaluate the importance of genes from the algebraic and information theory views, thereby selecting an optimal gene subset and improving classification precision. Third, Fisher score method is introduced into the proposed algorithm to preliminarily eliminate redundant genes to reduce the time cost of calculation and improve the performance of the algorithm. Finally, by comparing the experimental results of our algorithm with those of existing gene selection algorithms on ten gene datasets, it is proved that our algorithm can effectively improve the classification results for gene data.





Similar content being viewed by others
References
Liu KY, Yang XB, Yu HL, Fujita H, Chen XJ, Liu D (2020) Supervised information granulation strategy for attribute reduction. Int J Mach Learn Cybern 11(9):2149–2163. https://doi.org/10.1007/s13042-020-01107-5
Xu JC, Qu KL, Meng XR, Sun YH, Hou QC (2022) Feature selection based on multiview entropy measures in multiperspective rough set. Int J Intell Syst 37(10):7200–7234. https://doi.org/10.1002/int.22878
Sang BB, Chen HM, Yang L, Li TR, Xu WH (2022) Incremental feature selection using a conditional entropy based on fuzzy dominance neighborhood rough sets. IEEE Trans Fuzzy Syst 30(6):1683–1697. https://doi.org/10.1109/TFUZZ.2021.3064686
Qian WB, Dong P, Wang YL, Dai SM, Huang JT (2022) Local rough set-based feature selection for label distribution learning with incomplete labels. Int J Mach Learn Cybern 13(8):2345–2364. https://doi.org/10.1007/s13042-022-01528-4
Yang YY, Chen DG, Zhang X, Ji ZY, Zhang YJ (2022) Incremental feature selection by sample selection and feature-based accelerator. Appl Soft Comput. https://doi.org/10.1016/j.asoc.2022.108800
Chen Y, Liu KY, Song JJ, Fujita H, Yang XB, Qian YH (2020) Attribute group for attribute reduction. Inf Sci 535:64–80. https://doi.org/10.1016/j.ins.2020.05.010
Xu WH, Yuan KH, Li WT (2022) Dynamic updating approximations of local generalized multigranulation neighborhood rough set. Appl Intell 52(8):9148–9173. https://doi.org/10.1007/s10489-021-02861-x
Pawlak Z, Skowron A (2007) Rough sets: Some extensions. Inf Sci 177(1):28–40. https://doi.org/10.1016/j.ins.2006.06.006
Parthalain NM, Shen Q (2009) Exploring the boundary region of tolerance rough sets for feature selection. Pattern Recogn 42(5):655–667. https://doi.org/10.1016/j.patcog.2008.08.029
Sun L, Wang LY, Ding WP, Qian YH, Xu JC (2020) Neighborhood multi-granulation rough sets-based attribute reduction using Lebesgue and entropy measures in incomplete neighborhood decision systems. Knowl-Based Syst. https://doi.org/10.1016/j.knosys.2019.105373
Wang CZ, Huang Y, Shao MW, Hu QH, Chen DG (2020) Feature selection based on neighborhood self-information. IEEE Transactions on Cybernetics 50(9):4031–4042. https://doi.org/10.1109/TCYB.2019.2923430
Hu QH, Yu DR, Liu JF, Wu CX (2008) Neighborhood rough set based heterogeneous feature subset selection. Inf Sci 178(18):3577–3594. https://doi.org/10.1016/j.ins.2008.05.024
Sun L, Xu JC, Tian Y (2012) Feature selection using rough entropy-based uncertainty measures in incomplete decision systems. Knowl-Based Syst 36:206–216. https://doi.org/10.1016/j.knosys.2012.06.010
Sun L, Wang LY, Ding WP, Qian YH, Xu JC (2021) Feature selection using fuzzy neighborhood entropy-based uncertainty measures for fuzzy neighborhood multigranulation rough sets. IEEE Trans Fuzzy Syst 29(1):19–33. https://doi.org/10.1109/TFUZZ.2020.2989098
Wang CZ, Huang Y, Ding WP, Cao ZH (2021) Attribute reduction with fuzzy rough self-information measures. Inf Sci 549:68–86. https://doi.org/10.1016/j.ins.2020.11.021
Tsumoto S (2002) Accuracy and coverage in rough set rule induction. Int Conf Rough Sets CurrTrends Comput. https://doi.org/10.1007/3-540-45813-1_49
Sun L, Zhang XY, Qian YH, Xu JC, Zhang SG (2019) Feature selection using neighborhood entropy-based uncertainty measures for gene expression data classification. Inf Sci 502:18–41. https://doi.org/10.1016/j.ins.2019.05.072
Wong SKM, Ziarko W (1985) On optimal decision rules in decision tables. Bull Polish Acad Sci Math 33(11):693–696
Hu QH, Zhang L, Zhang D, Pan W, An S, Pedrycz W (2011) Measuring relevance between discrete and continuous features based on neighborhood mutual information. Expert Syst Appl 38(9):10737–10750. https://doi.org/10.1016/j.eswa.2011.01.023
Sun L, Zhang XY, Qian YH, Xu JC, Zhang SG, Tian Y (2019) Joint neighborhood entropy-based gene selection method with fisher score for tumor classification. Appl Intell 49(4):1245–1259. https://doi.org/10.1007/s10489-018-1320-1
Xu JC, Wang Y, Mu HY, Huang FZ (2019) Feature genes selection based on fuzzy neighborhood conditional entropy. J Intell Fuzzy Syst 36(1):117–126. https://doi.org/10.3233/JIFS-18100
Jiang ZH, Yang XB, Yu HL, Liu D, Wang PX, Qian YH (2019) Accelerator for multi-granularity attribute reduction. Knowl-Based Syst 177:145–158. https://doi.org/10.1016/j.knosys.2019.04.014
Fan J, Jiang YL, Liu Y (2017) Quick attribute reduction with generalized indiscernibility models. Inf Sci 397:15–36. https://doi.org/10.1016/j.ins.2017.02.032
Sun L, Zhang XY, Xu JC, Wang W, Liu RN (2018) A gene selection approach based on the fisher linear discriminant and the neighborhood rough set. Bioengineered 9(1):144–151. https://doi.org/10.1080/21655979.2017.1403678
Li WT, Xu WH, Zhang XY, Zhang J (2021) Updating approximations with dynamic objects based on local multigranulation rough sets in ordered information systems. Artif Intell Rev. https://doi.org/10.1007/s10462-021-10053-9
Miao DQ, Hu GR (1999) A heuristic algorithm for knowledge reduction. J Comput Res Dev 36(6):681–684
Wang GY, Yu H, Yang DC (2002) Decision table reduction based on conditional information entropy. Chin J Comput 25(7):759–766. https://doi.org/10.3321/j.issn:0254-4164.2002.07.013
Wu D, Guo SZ (2019) An improved Fisher Score feature selection method and its application. J Liaoning Tech Univ 38(5):472–479
Sun L, Zhang XY, Xu JC, Zhang SG (2019) An attribute reduction method using neighborhood entropy measures in neighborhood rough sets. Entropy. https://doi.org/10.3390/e21020155
Xu JC, Qu KL, Yang Y (2021) Feature selection combining information theory view and algebraic view in the neighborhood decision system. Entropy 23(6):704. https://doi.org/10.3390/e23060704
Chen XW, Xu WH (2022) Double-quantitative multigranulation rough fuzzy set based on logical operations in multi-source decision systems. Int J Mach Learn Cybern 13(4):1021–1048. https://doi.org/10.1007/s13042-021-01433-2
Shukla AK, Singh P, Vardhan M (2018) A hybrid gene selection method for microarray recognition. Biocybern Biomed Eng 38(4):975–991. https://doi.org/10.1016/j.bbe.2018.08.004
Ye CC, Pan JL, Jin Q (2019) An improved SSO algorithm for cyber-enabled tumor risk analysis based on gene selection. Future Gener Comput Syst 92:407–418. https://doi.org/10.1016/j.future.2018.10.008
Dong HB, Li T, Ding R, Sun J (2018) A novel hybrid genetic algorithm with granular information for feature selection and optimization. Appl Soft Comput 65:33–46. https://doi.org/10.1016/j.asoc.2017.12.048
Huang XJ, Zhang L, Wang BJ, Li FZ, Zhang Z (2018) Feature clustering based support vector machine recursive feature elimination for gene selection. Appl Intell 48(3):594–607. https://doi.org/10.1007/s10489-017-0992-2
Sun SQ, Peng QK, Zhang XK (2016) Global feature selection from microarray data using Lagrange multipliers. Knowl-Based Syst 110:267–274. https://doi.org/10.1016/j.knosys.2016.07.035
Sun L, Liu RN, Xu JC, Zhang SG, Tian Y (2018) An affinity propagation clustering method using hybrid kernel function with LLE. IEEE Access 6:68892–68909. https://doi.org/10.1109/ACCESS.2018.2880271
Xu FF, Miao DQ, Wei L (2009) Fuzzy-rough attribute reduction via mutual information with an application to cancer classification. Comput Math Appl 57(6):1010–1017. https://doi.org/10.1016/j.camwa.2008.10.027
Chen YM, Zhang ZJ, Zheng JZ, Ma Y, Xue Y (2017) Gene selection for tumor classification using neighborhood rough sets and entropy measures. J Biomed Inform 67:59–68. https://doi.org/10.1016/j.jbi.2017.02.007
Yang J, Liu YL, Feng CS, Zhu GQ (2016) Applying the Fisher score to identify Alzheimer’s disease-related genes. Genet Mol Res. https://doi.org/10.4238/gmr.15028798
Xu JC, Qu KL, Sun YH, Yang J (2022) Feature selection using self-information uncertainty measures in neighborhood information systems. Appl Intell. https://doi.org/10.1007/s10489-022-03760-5
Fan XD, Zhao WD, Wang CZ, Huang Y (2018) Attribute reduction based on max-decision neighborhood rough set model. Knowl-Based Syst 151:16–23. https://doi.org/10.1016/j.knosys.2018.03.015
Sun L, Xu JC, Wang W, Yin Y (2016) Locally linear embedding and neighborhood rough set-based gene selection for gene expression data classification. Genet Mol Res. https://doi.org/10.4238/gmr.15038990
Zhang W, Chen JJ (2018) Relief feature selection and parameter optimization for support vector machine based on mixed kernel function. J Mater Eng Perform 14(2):280–289. https://doi.org/10.23940/ijpe.18.02.p9.280289
Aziz R, Verma CK, Srivastava N (2016) A fuzzy based feature selection from independent component subspace for machine learning classification of microarray data. Genomics Data 8:4–15. https://doi.org/10.1016/j.gdata.2016.02.012
Apolloni J, Leguizamon G, Alba E (2016) Two hybrid wrapper-filter feature selection algorithms applied to high-dimensional microarray experiments. Appl Soft Comput 38:922–932. https://doi.org/10.1016/j.asoc.2015.10.037
Lu HJ, Chen JY, Yan K, Jin Q, Xue Y, Gao ZG (2017) A hybrid feature selection algorithm for gene expression data classification. Neurocomputing 256:56–62. https://doi.org/10.1016/j.neucom.2016.07.080
Li JT, Dong WP, Meng DY (2018) Grouped gene selection of cancer via adaptive sparse group lasso based on conditional mutual information. IEEE-ACM Trans Comput Biol Bioinform 15(6):2028–2038. https://doi.org/10.1109/TCBB.2017.2761871
Dunn QJ (1961) Multiple comparisons among means. J Am Stat Assoc 56(293):52–64. https://doi.org/10.1080/01621459.1961.10482090
Friedman M (1940) A comparison of alternative tests of significance for the problem of mrankings. Ann Math Stat 11(1):86–92. https://doi.org/10.1214/aoms/1177731944
Su ZG, Hu QH, Denoeux T (2021) A distributed rough evidential K-NN classifier: Integrating feature reduction and classification. IEEE Trans Fuzzy Syst 29(8):2322–2335. https://doi.org/10.1109/TFUZZ.2020.2998502
Xu WH, Yuan KH, Li WT, Ding WP (2022) An emerging fuzzy feature selection method using composite entropy-based uncertainty measure and data distribution. IEEE Trans Emerg Top Comput Intell. https://doi.org/10.1109/TETCI.2022.3171784
Acknowledgements
This work was supported in part by the National Natural Science Foundation of China under Grant (61976082, 62002103).
Author information
Authors and Affiliations
Contributions
JX: Conceptualization, Writing review and editing, Visualization, Project administration. KQ: Methodology, Software, Writing-original draft preparaton. QH: Formal analysis, Writing review and editing, Visualization. KQ: Writing review and editing, Visualization. XM: Formal analysis, Writing review and editing, Visualization. All authors have read and agreed to the published version of the manuscipt.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Appendix
Appendix
Proof of Proposition 1
From Eq. (2) and Eq. (8), we can get \(\left| {n_b^\delta \left( {{u_i}} \right) } \right| \ge \left| {n_c^\delta \left( {{u_i}} \right) } \right|\) and \({P_b}\left( D \right) \le {P_c}\left( D \right)\). According to Definition 4, \(N{H_\delta }\left( c \right) \ge N{H_\delta }\left( b \right)\) holds.
Proof of Property 1
According to Definition 6, \(N{H_\delta }\left( {D,c} \right) = N{H_\delta }\left( {D|c} \right) + N{H_\delta }\left( c \right)\) holds.
Proof of Proposition 2
Proof of Proposition 3
From Eq. (2), we know that \(n_b^\delta \left( {{u_i}} \right) \supseteq n_c^\delta \left( {{u_i}} \right)\), so \(n_b^\delta \left( {{u_i}} \right) \cap {\left[ {{u_i}} \right] _D} \supseteq n_c^\delta \left( {{u_i}} \right) \cap {\left[ {{u_i}} \right] _D}\), \(n_b^\delta \left( {{u_i}} \right) \cup {\left[ {{u_i}} \right] _D} \supseteq n_c^\delta \left( {{u_i}} \right) \cup {\left[ {{u_i}} \right] _D}\) and \({n_{\left( {b,D} \right) }}\left( {{u_i}} \right) \supseteq {n_{\left( {c,D} \right) }}\left( {{u_i}} \right)\). Thus, the numerical relationship between \(\frac{{{{\left| {n_b^\delta \left( {{u_i}} \right) \cap {{\left[ {{u_i}} \right] }_D}} \right| }^2}}}{{\left| {{n_{\left( {b,D} \right) }}\left( {{u_i}} \right) } \right| }}\) and \(\frac{{{{\left| {n_c^\delta \left( {{u_i}} \right) \cap {{\left[ {{u_i}} \right] }_D}} \right| }^2}}}{{\left| {{n_{\left( {c,D} \right) }}\left( {{u_i}} \right) } \right| }}\) is unknown, so the numerical relationship between \(- \frac{1}{{\left| U \right| }}\mathop \sum \limits _{i = 1}^{\left| U \right| } \mathrm{{log}}\left( {\frac{{{{\left| {n_b^\delta \left( {{u_i}} \right) \cap {{\left[ {{u_i}} \right] }_D}} \right| }^2}}}{{\left| U \right| \left| {{n_{\left( {b,D} \right) }}\left( {{u_i}} \right) } \right| }}} \right)\) and \(- \frac{1}{{\left| U \right| }}\mathop \sum \limits _{i = 1}^{\left| U \right| } \mathrm{{log}}\left( {\frac{{{{\left| {n_c^\delta \left( {{u_i}} \right) \cap {{\left[ {{u_i}} \right] }_D}} \right| }^2}}}{{\left| U \right| \left| {{n_{\left( {c,D} \right) }}\left( {{u_i}} \right) } \right| }}} \right)\) is not clear. From Eq. (8), \({P_b}\left( D \right) \le {P_c}\left( D \right)\) can be known. Therefore, the relation between \(- \frac{{{P_b}\left( D \right) }}{{\left| U \right| }}\mathop \sum \limits _{i = 1}^{\left| U \right| } \mathrm{{log}}\left( {\frac{{{{\left| {n_b^\delta \left( {{u_i}} \right) \cap {{\left[ {{u_i}} \right] }_D}} \right| }^2}}}{{\left| U \right| \left| {{n_{\left( {b,D} \right) }}\left( {{u_i}} \right) } \right| }}} \right)\) and \(- \frac{{{P_c}\left( D \right) }}{{\left| U \right| }}\mathop \sum \limits _{i = 1}^{\left| U \right| } \mathrm{{log}}\left( {\frac{{{{\left| {n_c^\delta \left( {{u_i}} \right) \cap {{\left[ {{u_i}} \right] }_D}} \right| }^2}}}{{\left| U \right| \left| {{n_{\left( {c,D} \right) }}\left( {{u_i}} \right) } \right| }}} \right)\) is not clear. According to Eq. (20), Proposition 3 holds.
Example
A Neighborhood decision system \(NS = \left( {U,C,D,\,\delta } \right)\) is shown below, where the universe \(U = \left\{ {{u_1},{u_2},{u_3},{u_4}} \right\}\); the conditional attribute set \(C = \left\{ {{c_1},{c_2},{c_3}} \right\}\); the decision attribute \(D = d\); the neighborhood radius \(\delta = 0.3\). Let initial gene subset \(c = \emptyset\), the base of log is 10, and \(P = 2\) in Eq. (1).
U | \({c_1}\) | \({c_2}\) | \({c_3}\) | d |
|---|---|---|---|---|
\({u_1}\) | 0.12 | 0.41 | 0.61 | Y |
\({u_2}\) | 0.21 | 0.15 | 0.14 | Y |
\({u_3}\) | 0.31 | 0.11 | 0.26 | N |
\({u_4}\) | 0.61 | 0.13 | 0.23 | N |
From Eq. (3), \({\left[ {{u_1}} \right] _D} = {\left[ {{u_2}} \right] _D} = \left\{ {{u_1},{u_2}} \right\}\), \({\left[ {{u_3}} \right] _D} = {\left[ {{u_4}} \right] _D} = \left\{ {{u_3},{u_4}} \right\}\).
From Eq. (1), when \(c = \left\{ {{c_1}} \right\}\), we know that \(D{F_{\left\{ {{c_1}} \right\} }}\left( {{u_1},\,{u_1}} \right) = 0 \le \delta\), \(D{F_{\left\{ {{c_1}} \right\} }}\left( {{u_1},\,{u_2}} \right) = 0.09 \le \delta\), \(D{F_{\left\{ {{c_1}} \right\} }}\left( {{u_1},\,{u_3}} \right) = 0.19 \le \delta\), \(D{F_{\left\{ {{c_1}} \right\} }}\left( {{u_1},\,{u_4}} \right) = 0.49 > \delta\), \(D{F_{\left\{ {{c_1}} \right\} }}\left( {{u_2},\,{u_2}} \right) = 0 \le \delta\), \(D{F_{\left\{ {{c_1}} \right\} }}\left( {{u_2},\,{u_3}} \right) = 0.1 \le \delta\), \(D{F_{\left\{ {{c_1}} \right\} }}\left( {{u_2},\,{u_4}} \right) = 0.4 > \delta\), \(D{F_{\left\{ {{c_1}} \right\} }}\left( {{u_3},\,{u_3}} \right) = 0 \le \delta\), \(D{F_{\left\{ {{c_1}} \right\} }}\left( {{u_3},\,{u_4}} \right) = 0.3 \le \delta\), \(D{F_{\left\{ {{c_1}} \right\} }}\left( {{u_4},\,{u_4}} \right) = 0 \le \delta\).
From Eq. (2), \(n_{\left\{ {{c_1}} \right\} }^\delta \left( {{u_1}} \right) = \left\{ {{u_1},{u_2},{u_3}} \right\}\), \(n_{\left\{ {{c_1}} \right\} }^\delta \left( {{u_2}} \right) = \left\{ {{u_1},{u_2},{u_3}} \right\}\), \(n_{\left\{ {{c_1}} \right\} }^\delta \left( {{u_3}} \right) = \left\{ {{u_1},{u_2},{u_3},{u_4}} \right\}\), \(n_{\left\{ {{c_1}} \right\} }^\delta \left( {{u_4}} \right) = \left\{ {{u_3},{u_4}} \right\}\).
From Eq. (14), \({n_{\left( {\left\{ {{c_1}} \right\} ,D} \right) }}\left( {{u_1}} \right) = n_{\left\{ {{c_1}} \right\} }^\delta \left( {{u_1}} \right) \cup {\left[ {{u_1}} \right] _D} = \left\{ {{u_1},{u_2},{u_3}} \right\}\), \({n_{\left( {\left\{ {{c_1}} \right\} ,D} \right) }}\left( {{u_2}} \right) = n_{\left\{ {{c_1}} \right\} }^\delta \left( {{u_2}} \right) \cup {\left[ {{u_2}} \right] _D} = \left\{ {{u_1},{u_2},{u_3}} \right\}\), \({n_{\left( {\left\{ {{c_1}} \right\} ,D} \right) }}\left( {{u_3}} \right) = n_{\left\{ {{c_1}} \right\} }^\delta \left( {{u_3}} \right) \cup {\left[ {{u_3}} \right] _D} = \left\{ {{u_1},{u_2},{u_3},{u_4}} \right\}\), \({n_{\left( {\left\{ {{c_1}} \right\} ,D} \right) }}\left( {{u_4}} \right) = n_{\left\{ {{c_1}} \right\} }^\delta \left( {{u_4}} \right) \cup {\left[ {{u_4}} \right] _D} = \left\{ {{u_3},{u_4}} \right\}\).
From Eq. (6), Eq. (7), and Eq. (8), \(\underline{{N_{\left\{ {{c_1}} \right\} }}} \left( D \right) = \,\left\{ {{u_4}} \right\}\), \(\overline{{N_{\left\{ {{c_1}} \right\} }}} \left( D \right) = \left\{ {{u_1},{u_2},{u_3},{u_4}} \right\}\), \({P_{\left\{ {{c_1}} \right\} }}\left( D \right) = \,\frac{{\left| {\underline{{N_{\left\{ {{c_1}} \right\} }}} \left( D \right) } \right| }}{{\left| {\overline{{N_{\left\{ {{c_1}} \right\} }}} \left( D \right) } \right| }} = \frac{1}{4}\).
From Eq. (20), \(N{H_\delta }\left( {D,\left\{ {{c_1}} \right\} } \right) = - \frac{{{P_{\left\{ {{c_1}} \right\} }}\left( D \right) }}{{\left| U \right| }}\mathop \sum \limits _{i = 1}^{\left| U \right| } \log \left( {\frac{{{{\left| {n_{\left\{ {{c_1}} \right\} }^\delta \left( {{u_i}} \right) \cap {{\left[ {{u_i}} \right] }_D}} \right| }^2}}}{{\left| U \right| \left| {{n_{\left( {\left\{ {{c_1}} \right\} ,D} \right) }}\left( {{u_i}} \right) } \right| }}} \right)\)
\(= - \frac{1/4}{4}\left( {\log \left( {\frac{{{2^2}}}{{4 \times 3}}} \right) + \log \left( {\frac{{{2^2}}}{{4 \times 3}}} \right) + \log \left( {\frac{{{2^2}}}{{4 \times 4}}} \right) + \log \left( {\frac{{{2^2}}}{{4 \times 2}}} \right) } \right) = 0.116\)
Similarly, \(N{H_\delta }\left( {D,\left\{ {{c_2}} \right\} } \right) = 0\), \(N{H_\delta }\left( {D,\left\{ {{c_3}} \right\} } \right) = 0.191\), \(N{H_\delta }\left( {D,\left\{ {{c_1},{c_2}} \right\} } \right) = 0.345\), \(N{H_\delta }\left( {D,\left\{ {{c_1},{c_3}} \right\} } \right) = 0.496\), \(N{H_\delta }\left( {D,\left\{ {{c_2},{c_3}} \right\} } \right) = 0.191\), \(N{H_\delta }\left( {D,\left\{ {{c_1},{c_2},{c_3}} \right\} } \right) = 0.496\).
From Eq. (21), when \(c = \emptyset\), \(Sig\left( {{c_2},\emptyset ,D} \right) = 0< Sig\left( {{c_1},\emptyset ,D} \right) = 0.116 < Sig\left( {{c_3},\emptyset ,D} \right) = 0.191\), so \({c_3}\) is added into c. Because \(Sig\left( {{c_2},\left\{ {{c_3}} \right\} ,D} \right) = 0 < Sig\left( {{c_1},\left\{ {{c_3}} \right\} ,D} \right) = 0.305\), \({c_1}\) is added into c. Because \(Sig\left( {{c_2},\left\{ {{c_1},{c_3}} \right\} ,D} \right) = 0\) satisfies the termination condition, \(c = \left\{ {{c_1},{c_3}} \right\}\) is an optimal gene subset.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Xu, J., Qu, K., Qu, K. et al. Feature selection using neighborhood uncertainty measures and Fisher score for gene expression data classification. Int. J. Mach. Learn. & Cyber. 14, 4011–4028 (2023). https://doi.org/10.1007/s13042-023-01878-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13042-023-01878-7