Abstract
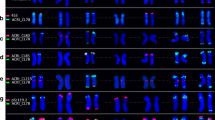
Chromosomes of Korean hexaploid wheat were investigated to compare the chromosomal karyotype for cytogenetic diversity. Chromosomal karyotyping was done with in situ hybridization using two types of simple sequence repeats (SSR)s, (AAG)5 and (AAC)5 labeled with tetramethyl-rhodamine-5-dUTP and fluorescein-12-dUPT as a fluorescence, respectively. The two SSRs as cytogenetic markers revealed that the cytogenetic characteristics of the wheat chromosomes were remarkably a B genome. In this study, the chromosomal karyotype of Keumkang, a Korean hexaploid wheat cultivar, was the A, B, and D genomes used as a cytogenetic reference. The expressed signals from the two SSRs showed a difference in the chromosomal karyotype of chromosome 1B among the Korean hexaploid wheat. The distribution pattern and the degree of condensation for the (AAG)5 and (AAC)5 signals on the short arm of chromosome 1B were different in the Korean hexaploid wheat shown in descending order: Keumkang > Joeun > Johan > Olgeuru. Olgeuru had a lower level of distribution and condensation for the two SSRs signals compared to the other Korean hexaploid wheat. In the A genome, chromosome 7A showed an unbalanced expression of the (AAG)5 signal on the distal region of the short and long arms in several Korean hexaploid wheat while Joeun, a Korean hexaploid wheat, showed a definite (AAG)5 signal on the distal region of each arm of chromosome 7A. Among the Korean hexaploid wheat, Shinmichal1, a Korean hexaploid waxy wheat, had a chromosome with a unique expression pattern for (AAG)5 and (AAC)5 compared the other Korean hexaploid wheat. Those cytogenetic differences identified in this study are useful as an indicator to improve the cytogenetic diversity in the Korean wheat breeding program.
Similar content being viewed by others
References
Arumuganathan K, Earle ED. 1991. Nuclear DNA content of some important plant species, Plant Mol. Bio. Rep. 9: 208–218
Badaeva ED, Keilwagen J, Knüpffer H, WaBermann L, Dedkova OS, Mitrofanova OP, Kovaleva ON, Liapunova OA, Pukhalskiy VA, Őzkan H, Graner A, Willcox G, Kilian B. 2015. Chromosomal passports provide new insights into diffusion of emmer wheat. PLOS ONE 10: e0128556
Choulet F, Wicker T, Rustenholz C, Paux E, Salse J, Leroy P, Schlub S, Le Paslier MC, Magdelenat G, Gonthier C, Couloux A, Budak H, Breen J, Pumphrey M, Liu S, Kong X, Jia J, Gut M, Brunel D, Anderson JA, Gill BS, Appels R, Keller B, Feuillet C. 2010. Megabase level sequencing reveals contrasted organization and evolution patterns of the wheat gene and transposable element spaces. The Plant Cell 22: 1686–1701
Cuadrado A, Cardoso A, Jouve N. 2008. Physical organization of simple sequence repeats (SSRs) in Triticeae: structural, functional and evolutionary implications. Cytogenet. Genome Res. 120: 210–219
Cuadrado A, Schwarzacher T. 1998. The chromosomal organization of simple sequence repeats in wheat and rye genomes. Chromosoma 107: 587–594
Cuadrado A, Schwarzacher T, Jouve N. 2000. Identification of different chromatin classes in wheat using in situ hybridization with simple sequence repeat oligonucleotides. Theor. Appl. Genet. 101: 711–717
Dvorák J, McGuire PE. Nonstructural chromosome differentiation among wheat cultivars, with special reference to differentiation of chromosomes in related species. Genetics 1981: 391–414
Gill BS, Friebe B, Endo TR. 1991. Standard karyotype and nomenclature system for description of chromosome bands and structural aberrations in wheat (Triticum aestivum). Genome 34: 830–839
Gill BS, Kimber G. 1974. Giemsa C–banding and the evolution of wheat. Proc. Natl. Acad. Sci. 71: 4086–4090
Huang S, Sirikhachornkit A, Faris JD, Su X, Gill BS, Haselkorn R, Gornicki P. 2002a. Phylogenetic analysis of the acetyl–CoA carboxylase and 3–phosphoglycerate kinase loci in wheat and other grasses. Plant Mol. Biol. 48: 805–820
Huang S, Sirikhachornkit A, Su X, Faris J, Gill B, Haselkorn R, Gornicki P. 2002b. Genes encoding plastid acetyl–CoA carboxylase and 3–phosphoglycerate kinase of the Triticum/Aegilops complex and the evolutionary history of polyploid wheat. Proc. Natl. Acad. Sci. 99: 8133–8138
Kohno K, Takahashi H, Endo TR, Matsuo H, Shiwaku K, Morita E. 2016. Characterization of a hypoallergenic wheat line lacking ω–5 gliadin. Allergo. Int. 65: 400–405
McFadden ES, Sears ER. 1946. The origin of Triticum spelta and its free threshing hexaploid relatives. J. Hered. 37: 81–107
Mirzaghaderi G, Houben A, Badaeva ED. 2014. Molecularcytogenetic analysis of Aegilops triuncialis and identification of its chromosomes in the background of wheat. Mol. Cytogenetics 7: 91
Naranjo T, Roca P, Goicoechea PG, Giraldez R. 1987. Arm homology of wheat and rye chromosomes. Genome 29: 873–882
Sourdille P, Tavaud M, Charmet G, Bernard M. 2001. Transferability of wheat microsatellites to diploid Triticeae species carrying the A, B and D genomes. Theor. Appl. Genet. 103: 346–352
Talbert LE, Blake NK, Storlie EW, Lavin M. 1009. Variability in wheat based on low–copy DNA sequence comparisons. Genome 38: 951–957
Tang Z, Li M, Chen L, Wang Y, Ren Z, Fu S. 2014. New types of wheat chromosomal structural variations in derivatives of wheat–rye hybrids. PLOS ONE 9: e110282
Zimin AV, Puiu D, Hall R, Kingan S, Clavijo BJ, Salzberg SL. 2017. The first near–complete assembly of the hexaploid bread wheat genome, Triticum aestivum. GigaSci. 6: 1–7
Zhang H, Li G, Li D, Gao D, Zhang J, Yang E, Yang Z. 2015. Molecular and cytogenetic characterization of new wheat–Dasypyrum breviaristatum derivatives with post–harvest re–growth habit. Genes 6: 1242–1255
Zheng JS, Sun CZ, Zhang SN, Hou XI, Bonnema G. 2016. Cytogenetic diversity of simple sequences repeats in morphotypes of Brassica rapa ssp. chinensis. Frontiers in Plant Sci. 7: 1049
Author information
Authors and Affiliations
Corresponding authors
Rights and permissions
About this article
Cite this article
Cho, SW., Kang, SW., Kang, TG. et al. Cytogenetic Diversity of Korean Hexaploid Wheat (Triticum aestivum L.) with Simple Sequence Repeats (SSRs) by Fluorescence In Situ Hybridization. J. Crop Sci. Biotechnol. 21, 491–497 (2018). https://doi.org/10.1007/s12892-018-0245-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12892-018-0245-0