Abstract
The polar bear (Ursus maritimus) is a species particularly vulnerable to the effects of climate change. As the climate warms, polar bears will be forced to move to more suitable habitats which are likely to shrink, adapt to the new conditions, or decline in population size. However, the genomic diversity within and among all 19 subpopulations of polar bears, and therefore their adaptive potential, is currently unknown. In addition, warmer climates are likely to result in more frequent contact between polar bears and grizzly bears (U. arctos), with which they can hybridize. Here we describe the development, quality control, and application of the Ursus maritimus V2 SNP chip. This 8 K SNP chip contains loci explicitly selected to assess both RAD-derived and transcriptome-derived loci, as well as SNPs to detect hybridization between species. A total of 7,239 loci (90.3% of those printed) were successfully genotyped, with over 99% genotype concordance for individuals typed in duplicate on this chip, and between individuals typed here and on the Ursus maritimus V1 SNP chip. Using simulations, we demonstrate that the markers have high accuracy and efficiency to detect hybridization and backcrosses between polar bears and grizzly bears. However, empirical analysis of 371 polar bears, 440 grizzly bears, and 8 known hybrids found no novel instances of recent hybridization. The Ursus maritimus V2 SNP chip provides a powerful tool for monitoring the adaptive potential of this species along with assessing population structure, quantitative genomics, and hybridization in polar bears.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In the face of global climate change, species have a limited number of possible responses: move to more favorable habitats, adapt to the new conditions, or suffer local extinction. For species residing in environments such as the Arctic, potential for movement is often limited, and therefore adaptation may be key to prevent declines. The relatively new concept of “evolutionarily enlightened management” aims to both protect species and their ability to evolve to changing environmental conditions (Ashley et al. 2003; Sgrò et al. 2011; Cook and Sgrò 2019). As such, it becomes critical for conservationists and managers to assess the adaptive potential of populations and species (Harrisson et al. 2014). Adaptive potential can be broken down into two nonexclusive components: (1) overall genetic diversity, considered the raw material for evolution; and (2) functional genetic diversity, the variation that underlies differences in traits, including the ability to cope with specific environmental conditions or use alternative habitats. Though both components are important for long-term persistence of species, they are rarely assessed simultaneously. In addition, for conservation to be maximally effective, managers need to know how genetic variation is distributed within and among populations (Funk et al. 2012).
The polar bear (Ursus maritimus) is a species particularly vulnerable to the effects of climate change, which has in some areas resulted in declines in individual polar bear fitness and is threatening population viability and persistence at a broader scale (Derocher et al. 2004; Stirling and Derocher 2012; Laidre et al. 2018, 2020). Changes in sea ice conditions will likely influence the movement of individuals and genetic structure of polar bear populations, potentially altering the distribution of local adaptive variation (Stirling and Derocher 2012; Laidre et al. 2018, 2022). Consequently, the long-term conservation of polar bears can be informed by an understanding of adaptive potential and genetic diversity across their range, especially in areas of the Queen Elizabeth Islands and Norwegian Bay within the Canadian Arctic Archipelago for which we have little genetic information (Hamilton et al. 2014; Molnár et al. 2020). Local adaptive variation may be further impacted by hybridization between polar bears and grizzly bears (U. arctos) as the latter shift northward (Rockwell et al. 2008) and polar bear behavioral patterns are altered with changing environmental conditions. Contemporary hybridization has already been observed in the western Canadian Arctic, where a polar bear female mated with two grizzly bear males who also sired the subsequent backcrossed generation (Pongracz et al. 2017). However, both recent and historical hybridization events indicate unidirectional movement of polar bear genes into grizzly bears, suggesting strong selection against grizzly bear genes in polar bears (Hailer 2015) or sex-biased directional hybridization (Pongracz et al. 2017).
Here, we describe the development, quality control, and application of a new custom-made genomic tool: the Ursus maritimus V2 SNP chip. Single nucleotide polymorphisms (SNPs) have rapidly become the marker of choice for molecular ecology, evolutionary biology, and conservation genomic studies because they have several advantages over other types of genetic markers (Morin et al. 2004; Helyar et al. 2011). SNPs are the most abundant form of genetic variation occurring in both neutral and functional regions of the genome.
With advances in high-throughput sequencing technologies, along with their decreasing costs, several avenues have opened for SNP discovery and genotyping in nearly any organism (Glenn 2011; van Dijk et al. 2018). The ultimate form of marker discovery and genotyping would be whole genome sequencing that theoretically would enable detection of all of the genetic variation within an organism, including nucleotide changes and structural variations such as insertion/deletions or inversions (Ellegren 2014; Fuentes-Pardo and Ruzzante 2017). However, whole genome sequencing remains relatively expensive, bioinformatically challenging, and likely unnecessary for many applications especially with respect to conservation (McMahon et al. 2014).
In contrast, a variety of reduced representation sequencing methods offer simultaneous marker discovery and genotyping for more targeted portions of the genome (Peterson et al. 2012; Campbell et al. 2018). These methods have found a wide range of applications in molecular ecology, phylogenetics, and conservation genomics (Andrews et al. 2016; Suchan et al. 2017). However, research has shown that marker accuracy is dependent on bioinformatic pipelines and filtering options employed, with no widely agreed upon standards (Davey et al. 2013; Gautier et al. 2013; Shafer et al. 2017; Díaz-Arce and Rodríguez-Ezpeleta 2019). Despite new methods such as hybrid capture (Jones and Good 2016), amplicon sequencing (Meek and Larson 2019), or size-selection free library preparations (MacDonald et al. 2020), the variability in accuracy of reduced-representation sequencing raises concerns about its repeatability across laboratories or even library preparations.
As an alternative, SNP chips offer a means for rapid and reliable genotyping. Based on microarray technologies, SNP chips generate hundreds to millions of genotypes at individual loci. SNP chips have long been used in many domestic organisms (e.g. McCue et al. 2012; Song et al. 2013; Kijas et al. 2014) and have been developed for a number of wild species (e.g. Hagen et al. 2013; Kawakami et al. 2014; Judkins et al. 2020). Although loci on a chip must be designated a priori and cannot be altered once developed, SNP chips have the benefits of high genotyping accuracy, low levels of missing data, and easy application across laboratories (Oliphant et al. 2002; Shen et al. 2005; Helyar et al. 2011). This latter characteristic is especially beneficial for situations where there are multiple groups working on similar questions across a species’ range, which is the case with polar bears, and it can facilitate more integrated range-wide conservation efforts. These characteristics have led to the continued use and development of SNP chips (e.g. Judkins et al. 2020; Humble et al. 2020; Duntsch et al. 2020; Lee et al. 2021).
Building upon a previous version of this technology, Ursus maritimus V1 (Malenfant et al. 2015), the Ursus maritimus V2 SNP chip was specifically designed to assess both overall and functional genetic diversity across the polar bear range, as well as to distinguish between polar bears and grizzly bears and assess hybrids. We define hybrids to include both F1 hybrids and other backcrossed generations. Our objective was to address three questions:
-
1)
What is the rate of marker conversion and accuracy of the Ursus maritimus V2 SNP chip?
-
2)
What is the power of the new set of markers to detect hybrid individuals?
-
3)
What is the frequency of contemporary or past hybridization events?
Methods and materials
Design of the SNP chip and marker conversion
The starting point for this work was the Ursus maritimus V1 SNP chip developed by Malenfant et al. (2015). Briefly, this chip contained loci derived from two main sources: (1) a transcriptome assembly from two tissue types (blood and fat) of 10 polar bear individuals (Genetic Resources Development Consortium et al. 2014), allowing assessment of genetic diversity likely enriched for functional variants, and (2) a de novo restriction-site associated DNA (RAD) assembly based on 38 polar bears from across their range, allowing assessment of primarily neutral genetic diversity (Malenfant et al. 2015).
The Ursus maritimus V2 SNP chip builds on this work by incorporating additional loci, primarily focusing on those that would facilitate species identification and characterization of hybridization between polar bears and grizzly bears. Specifically, in addition to all variable RAD and transcriptomic loci from the first chip, the new chip contained markers from the following sources: replacement transcriptomic loci (i.e., SNPs in a gene included on the first chip but for which the original SNP was invariant or otherwise failed), previously unused transcriptomic loci (i.e., those in genes not included in the first chip), and species-discriminating loci. These species-discriminating loci were extracted from four sources: (1) fixed differences from Cronin et al. (2014); (2) diagnostic loci from Liu et al. (2014); (3) Y-chromosome markers from Bidon et al. (2014); and (4) mitochondrial SNPs from Miller et al. (2012b). Note that in the set of loci from Liu et al. (2014), preference was given to markers showing complete fixation of genotypes between species, but in order to increase the number of candidates, we included some loci that had one alternate allele shared with polar bears (among the 10 grizzly bear individuals considered) on the chip. We included loci from the Y-chromosome and mitochondrial genome to assess the direction of hybridization. The mitochondrial loci were selected such that we could distinguish polar bears from grizzly bears, as well as grizzly bears from the Admiralty, Baranof, and Chichagof (ABC) Islands of Alaska compared to other populations of grizzly bears because of the complex demographic history of grizzly bears on the ABC Islands (Cahill et al. 2013, 2015; Bidon et al. 2014; Hassanin 2015). A total of 9,003 loci were submitted for Illumina Infinium BeadChip (Illumina, San Diego, CA, United States of America [USA]) design, were marked as Infinium II SNPs, and were tiled in a single direction. Twenty-eight loci were identified as duplicate in the design (either by name or by sequence) and one of each pair was removed from the final design. The final printed chip contained 8,015 loci in total.
Sample collection, DNA extraction, and genotyping
Samples of polar bears, grizzly bears, and known hybrids were collected between 1975 and 2015 (Table S1) from several long-term monitoring projects, capture–recapture studies, defense of life and property kills, and subsistence harvests operated by Environment and Climate Change Canada, Government of the Northwest Territories (Environment and Climate Change), and the Greenland Institute of Natural Resources and collaborators. All hybrid individuals were captured in the Northwest Territories (NWT) as described in Pongracz et al. (2017). Capture and handling protocols were consistent with the animal care guidelines set out for each project and approved by local Animal/Wildlife Care Committees including: the Canadian Council on Animal Care and Animal Care Committees of Environment and Climate Change Canada and Government of the Northwest Territories; Greenland Ministry for Hunting, Fisheries, and Agriculture, Greenland Ministry of the Environment, the Greenland veterinary authorities; as well as the U.S. Geological Survey Alaska Science Center Institutional Animal Care and Use Committee.
We genotyped blood and skin samples from 401 polar bears, 450 grizzly bears, and 8 known hybrids (Fig. 1, Table S1). Genotyping was performed by Delta Genomics (now Neogen; Edmonton AB, Canada [CA]). Specifically, we genotyped polar bears from collections in the Northern Beaufort Sea (N = 135), Southern Beaufort Sea (N = 112), based on the historical boundaries at the time of sample collection, Viscount Melville Sound (N = 124; including the mother of the known hybrid individuals), and East Greenland (N = 30) populations (Pongracz et al. 2017). The sampled grizzly bears mainly came from collections in the NWT (N = 421; including a father of some of the known hybrid individuals (Pongracz et al. 2017), though also included samples from Alberta, Canada (N = 20) and the ABC Islands of Alaska, USA (N = 9). We focused on these regions as they are nearest to areas where contemporary hybridization has been observed (Pongracz et al. 2017), except for East Greenland and Alberta, which represent allopatric populations that were used in simulations of hybridization (see below).
DNA was extracted from all samples using DNeasy Blood & Tissue Kits (Qiagen, Hilden, Germany). Whole-genome amplification, BeadChip hybridization, primer extension and staining were conducted according to Illumina’s recommended protocols (Illumina 2009). SNPs were genotyped using GenomeStudio 2011.1 (Genotyping Module 1.9; Illumina) using the automated clustering feature (default settings) after the removal of low‐quality samples (i.e., genotyping success rate < 0.9). Clustering patterns for all SNPs were manually examined, and SNPs were removed if they were monomorphic, had low call rates (< 0.99 for autosomal loci, < 0.50 for Y linked loci), or had unusual clustering patterns. Genotypes were then exported to PLINK formatted .ped and .map files with further manipulation of these files done with PLINK version 1.90b3 (Purcell et al. 2007; Chang et al. 2015) or PGDSpider version 2.0.5.1 (Lischer and Excoffier 2011).
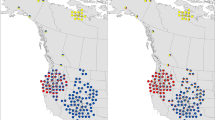
Map of study area and distribution of sampled individuals with known locations (N = 338 polar bears, 300 grizzly bears, and 8 hybrids [Pongracz et al. 2017]). Polar bears (PB) are depicted by the orange pluses, grizzly bears (GB) by the green triangles, and hybrids by the blue squares. Inset shows the main figure map placed in the context of the broader geographic area
Genotype concordance and genetic diversity
A total of 8 polar bear samples were genotyped twice on the Ursus maritimus V2 SNP chip, while N = 33 polar bear samples were genotyped on both the Ursus maritimus V1 and Ursus maritimus V2 SNP chips. This allowed us to measure concordance both within the newly developed chip and for loci held in common between the two typing systems (N = 4,723). Concordance was measured as the percentage of genotypes that match between samples typed in duplicate after discarding loci that did not successfully genotype in either sample.
Genetic diversity of markers was considered separately for each category of marker (RAD-derived, transcriptomic, and species-diagnostic) and within each species. Specifically, we considered the number of loci that were variable (minor allele frequency [MAF] > 0.01 and genotyped in at least 90% of samples), and the average MAF of those loci. MAF was calculated in PLINK with all individuals considered to be founders.
Hybridization power analysis
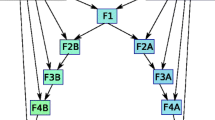
We used a simulation approach to examine the power of the new markers to detect hybrids and assign individuals to different generations of backcrosses. For this, two sets of reference samples were considered. The first set consisted of two parapatric populations with no expectation of recent gene flow between the species. The second set consisted of two sympatric reference populations which may have reduced differentiation due to a history of interbreeding. For the parapatric scenario, we used grizzly bears from Alberta (N = 20) and polar bears from East Greenland (N = 30) as the parental populations. For the sympatric scenario, both polar bear and grizzly bear samples came from close to Viscount Melville Sound (N = 15 GB and N = 30 PB), near the region where the two species are known to have encountered each other and interbred (Pongracz et al. 2017). These reference samples were used to simulate known hybrid generations via HybridLab version 1.0 (Nielsen et al. 2006). Specifically, we generated 50 individuals from each of the following hybrid categories: F1, F2 (mating between F1s), and 5 generations of F1 backcrosses to each of the parental species. Due to the computational limitations of NewHybrids (Anderson and Thompson 2022), the set of loci considered was restricted to the species-diagnostic SNPs from Cronin et al. (2014) and the fixed differences from Liu et al. (2014) (N = 287 loci total).
We assessed each simulated dataset with two assignment programs: NewHybrids version 1.1 (Anderson and Thompson 2002) and STRUCTURE version 2.3.4 (Pritchard et al. 2000; Falush et al. 2003). NewHybrids places individuals into pre-set ancestry categories (e.g. F1, or first-generation backcross to PB) and assigns a probability to that assignment. For the NewHybrids analyses, the program was run with Jeffreys prior using 100,000 burn-in iterations followed by 500,000 Markov chain Monte Carlo (MCMC) iterations. Analyses were repeated three times per dataset to ensure convergence in assignment among the runs, which was tested using CLUMPAK (Kopelman et al. 2015). Following Wringe et al. (2017), the power of the NewHybrids analyses was assessed for efficiency (i.e., the ability to assign simulated individuals to the correct hybrid category) and accuracy (i.e., the number of individuals correctly assigned to a hybrid category over the total number of individuals assigned to that hybrid category). Both efficiency and accuracy were assessed across a range of probability threshold values. For STRUCTURE analyses, we ran the program with 100,000 iterations, after a 50,000 burn-in period using an admixture model with correlated allele frequencies among populations and all other parameters remaining at default. We enforced only two populations (K) with 10 replicates for each sample set and results were processed with CLUMPAK to determine average ancestry coefficients (q-values) across runs. The power of the STRUCTURE analyses was assessed as the mean and standard deviation of q-values for each hybrid category.
Search for interspecific hybrids
We searched for evidence of contemporary and historical hybridization using 440 grizzly bears and 371 polar bears, excluding those from East Greenland. We also included the 8 known hybrid individuals. For these analyses we considered species-diagnostic loci with MAF > 0.01 across all samples and genotyped in > 90% of individuals (N = 1,470 loci). As this set of loci included 18 mitochondrial SNPs, but no Y-chromosome loci which were removed during the MAF filtering, we conducted the analyses below twice: once excluding the mitochondrial loci and once including all loci. When mitochondrial loci were included in the analysis, they were coded as homozygous diploids.
We searched for hybrids using STRUCTURE with the same run parameters as above. However, here we explored K values between one and four with 10 repetitions of each K. Results were processed with CLUMPAK and optimal K was determined by looking at both mean ln(ProbK) as well as the delta K method (Evanno et al. 2005; Gilbert et al. 2012; Janes et al. 2017). Finally, we conducted a principal component analysis (PCA) with the R package LEA version 2.6.0 (Frichot and François 2015) using R version 3.6.0 (R Core Team 2019).
Results
Genotype concordance and genetic diversity
In total, 7,239 loci (90.3%) were successfully genotyped on the Ursus maritimus V2 SNP chip. Across the 8 polar bear samples typed in duplicate on the Ursus maritimus V2 SNP chip, concordance ranged from 99.78 to 100% (average 99.96%). Results were similar when considering the 33 polar bear samples in common between the Ursus maritimus V1 SNP chip and the Ursus maritimus V2 SNP chip. We assessed genetic variability across 400 polar bears and 449 grizzly bears, excluding the known hybrids and their assigned parents (Table 1). Across all samples individual call rates averaged 99.48% (SD = 0.46%), with polar bears having a slightly higher average than grizzly bears (99.91% ± 0.20% versus 99.09% ± 0.22%, respectively). At the 4,723 loci in common between the V1 and V2 chips, concordance ranged from 99.23 to 99.60% (average 99.44%). Of the 2,730 transcriptomic loci, 2,411 and 309 were variable in polar bears and grizzly bears, respectively. Of the 3,026 RAD-derived loci, 2,625 and 416 were variable in polar bears and grizzly bears, respectively. Of the 1,483 species-diagnostic loci, 267 and 916 were variable in polar bears and grizzly bears, respectively. Finally, 1,936 and 5,598 loci were fixed in polar bears and grizzly bears, respectively.
Genetic diversity measures varied among the marker categories and between species. For transcriptome-derived loci, the number of variable loci and average MAF were higher for polar bears than in grizzly bears (Table 1). For species-diagnostic loci, grizzly bears had more variable markers, but average MAF was higher in polar bears for the variable loci; and for RAD-derived loci, there were more loci polymorphic in polar bears compared to grizzly bears, although MAF of the variable loci were the same. Average MAF was higher for loci that had one shared allele between species in the ascertainment panel compared to those showing completely fixed differences. Specifically, average MAF ± SD for grizzly bears was 0.09 ± 0.11 versus 0.07 ± 0.11 for shared and fixed loci, respectively. In polar bears, MAF values were 0.05 ± 0.11 versus 0.04 ± 0.10.
Hybridization power analysis
The three replicate NewHybrids runs produced highly consistent results within both the sympatric and parapatric reference panels. Efficiency and accuracy values for each hybrid category were highly correlated between the two reference panels (r = 0.99 and 0.73, respectively). Across all hybrid categories, accuracy was slightly higher for simulations based on the parapatric reference populations, whereas there was no trend for efficiency. Efficiency was high for assignment to the parental populations, F1, F2, and first-generation backcrosses regardless of threshold probabilities (Fig. 2). Efficiency was lower for advanced generation backcrosses and declined with increasing probability thresholds. Accuracy was high for F1, F2, as well as first- and second-generation backcrosses regardless of threshold probabilities. Advanced generation backcrosses had lower accuracy, but these increased with increasing threshold probabilities. Accuracy of assignment to parental populations was lower than even some advanced generation backcrosses, especially at lower probability thresholds, due to misassignment of fourth- and fifth-generation backcrosses to these parental categories.
Plots of the accuracy (top row) and efficiency (bottom row) of assignment for each of the 14 hybrid categories as a function of probability threshold values. In hybrid category naming, backcrosses are indicated with the parental population followed by “BC” and the generation number, e.g. PB_BC5 is a fifth-generation backcross to a polar bear parent. Values involving grizzly bears (GB) are displayed with dashed lines, those involving polar bears (PB) are displayed with solid lines
In the STRUCTURE analyses, q-values reflected anticipated crosses and could distinguish early generation backcrosses (BC1–BC3), possibly BC4, but not F1 from F2 or BC5 from parental populations. Use of either the parapatric or sympatric parental populations produced nearly identical q-values except for grizzly bear backcrosses where the parapatric reference populations had higher average q-values for polar bear ancestry (Fig. 3).
Histogram showing distribution of q-values to polar bear (PB) ancestry (0 being fully grizzly bear (GB), 1 being fully polar bear) for each of the 50 simulated individuals among the 14 hybrid categories. In hybrid category naming, backcrosses are indicated with the parental population followed by “BC” and the generation number, e.g. PB_BC5 is a fifth-generation backcross to a polar bear parent
Search for interspecific hybrids
STRUCTURE analyses were nearly identical between datasets with and without mitochondrial loci. In both cases, K = 2 was heavily favoured by both mean ln(ProbK) as well as the delta K method with all 10 runs converging on a single solution (Tables S2 and S3). At K = 2, polar bears were differentiated from grizzly bears, and all known hybrid individuals were identified (mean proportion of genome derived from polar bear = 0.438; range 0.23 to 0.504; Fig. 4 and Figure S1). No novel hybrids were seen, and no grizzly bear ancestry was found in the polar bear samples. It appears that some polar bear ancestry was present in grizzly bears from the ABC Islands and additional individuals from Alberta (mean ± SD proportion of genome derived from PB = 0.022 ± 0.005).
Ancestry bar plot from STRUCTURE (Pritchard et al. 2000) of K = 2 clusters for 371 polar bears (Ursus maritimus; PB), 440 grizzly bears (Ursus arctos; GB), including those from the Admiralty, Baranof, and Chichagof (ABC) Islands of Alaska, and 8 known hybrids that were genotyped at 1,452 species-discriminating loci (excluding mitochondrial SNPs). Each individual is represented as a vertical bar, with the proportion of colors representing their genetic assignment to the clusters
Both species were also well resolved from one another in the PCA analysis (Fig. 5 and Figure S2) where the first principal component (PC) axis accounted for > 80% of variation in the dataset. The two species were clearly differentiated along this axis, and the known hybrids clustered between the two parental species. Within the hybrid cluster, individuals formed two groups that corresponded to previous microsatellite work (Pongracz et al. 2017), one group with individuals that were assigned as F1s (N = 4) and one with individuals that were assigned as grizzly bear backcrosses (N = 4). Interestingly, there was a difference across PC2 between datasets with and without the mitochondrial loci. Without inclusion of the mitochondrial loci all grizzly bear samples formed a single large cluster (Fig. 5), whereas inclusion of the mitochondrial loci led to separation of ABC Islands grizzly bears from the other grizzly bear populations (Figure S2).
PCA plot for the 819 samples genotyped at 1,452 species-discriminating loci (excluding mitochondrial SNPs). Shown are PC1 vs. PC2 with each sample represented as a point. Polar bears (PB) are depicted by the orange pluses, grizzly bears (GB) by the green triangles, and hybrids by the blue squares. Grizzly bears from the Admiralty, Baranof, and Chichagof (ABC) Islands of Alaska are shown as blue circles
Discussion
The Ursus maritimus V2 SNP chip represents a novel and important genomic tool for assessing and monitoring the genetic diversity and adaptive potential of polar bears. We observed high marker conversion and retention of polymorphic loci with an overall total of 7,239 loci (90.3% of those printed) successfully genotyped, as well as 5,036 of the 5,756 RAD and transcriptome-derived loci (87.5%) polymorphic in polar bears. This level of conversion is higher than other SNP chips developed for wild species, including salmon (Salmo salar, 64.5%; Karlsson et al. 2011) and house sparrows (Passer domesticus, 74.1%; Hagen et al. 2013), though on par with one developed for Ficedula flycatchers (91.2%; Kawakami et al. 2014). Such high conversion is encouraging, and we expect additional loci to show variability when the chip is applied to additional populations of polar bears. Higher levels of marker conversion and variability in RAD and transcriptomic SNPs in polar bears compared to grizzly bears is to be expected because those sets of loci were discovered using only polar bear samples; therefore, conversion to grizzly bears will be restricted to relatively rare cases of shared ancestral polymorphisms (Miller et al. 2012a). Encouragingly, the species-diagnostic loci had low variability in polar bears. The fact that 61.8% of the species-diagnostic loci showed variability in grizzly bears may be the result of allowing rare, shared alleles in the ascertainment panel of grizzly bears when selecting loci from the work of Liu et al. (2014). Alternatively, such variability may be the result of ancient introgression from polar bears alleles that have dispersed into the grizzly bear range (Cahill et al. 2015; Hailer 2015). Our finding that average MAF was higher for loci that had one shared allele between species in the ascertainment panel than those showing completely fixed differences is consistent with both hypotheses.
Extremely high concordance was observed both for samples typed in duplicate within the Ursus maritimus V2 SNP chip and when comparing samples genotyped on both Ursus maritimus chips. Such high concordance rates and low levels of missing data are key benefits of SNP chips (Oliphant et al. 2002; Shen et al. 2005; Helyar et al. 2011), especially relative to other methods of genotyping where missing data and data processing can be much larger issues (Davey et al. 2013; Gautier et al. 2013; Shafer et al. 2017). Concordance of genotypes between the Ursus maritimus V1 and V2 SNP chips is key for analyses necessitating the combined analyses of the newly generated genotypes with previous ones (e.g., pedigree construction or quantitative genetic analyses; Malenfant 2016; Malenfant et al. 2018).
The simulations of hybridization and backcrossing between species demonstrated that the markers used here have extremely high power to detect early generation hybrids and advanced generation hybrids up to third-generation backcrosses. This was true regardless of whether the simulations were based on sympatric or parapatric parental populations. Note that these simulations were based on a reduced number of loci (due to computational limitations of NewHybrids), focusing on those loci which showed fixed differences in the ascertainment panel from Cronin et al. (2014) and Lui et al. (2014). We cannot rule out that analysis of the full dataset which includes loci with rare, shared polymorphisms between species may decrease observed differentiation levels between the species, making detection of advanced generation hybrids more difficult. Additionally, long-distance immigrants that carry rare, population-specific alleles from highly diverged populations can be misidentified as hybrids when samples are obtained from a small area of the species range (Harmoinen et al. 2021). However, we believe that additional power is likely to be gained when using the full complement of species-diagnostic loci on the Ursus maritimus V2 SNP chip which will ultimately improve hybrid discovery.
Despite the high resolving power of our SNP set, no additional early generation hybrids between species were found beyond the eight previously known. The q-values of these hybrids reflected their expected ancestry, either F1 or backcross to grizzly bears (Pongracz et al. 2017), which again speaks to the accuracy and resolving power of the loci used. Lack of additional early generation hybrid individuals suggests that contemporary hybridization between 1975 and 2015 is rare despite increased sightings of grizzly bears within the range of polar bears in Canada (Doupé et al. 2007; Pongracz et al. 2017; Clark et al. 2018). This observation is consistent with the results of Pongracz et al. (2017) who found that all of the F1 hybrid individuals could be traced to a single female polar bear. Moreover, the absence of even low levels of cross-species ancestry in polar bears implies that there were no ancestral hybridization events that then backcrossed into the polar bear gene pool.
The signal of polar bear ancestry in grizzly bears from the ABC Islands is interesting given the complex demographic history of polar bears and grizzly bears from this region. Previous work has suggested that ABC Island grizzly bears are descended from a polar bear population that became isolated during the Pleistocene and over time received extensive male-biased gene flow from mainland grizzly bears such that the majority of the genome is from grizzly bears but polar bear alleles are still present (Cahill et al. 2013, 2015; Bidon et al. 2014; Hassanin 2015; Wang et al. 2022). Although it was initially thought that gene flow from polar bears into grizzly bears was restricted to the ABC Islands, whole genome sequencing has found polar bear ancestry in grizzly bears from mainland Alaska and from a population in Russia (Cahill et al. 2015, 2018). Furthermore, analyses of an ancient polar bear genome have highlighted additional polar bear ancestry in grizzly bears that is not seen from modern genomes alone (Wang et al. 2022). Our results showed polar bear ancestry in grizzly bears from Alberta, Canada, which may support an even larger geographical extent for ancient introgression of polar bear alleles into grizzly bears (Hailer 2015). However, caution is warranted because other grizzly bear populations more proximate to the range of polar bears do not show the same presence of low levels of ancestry.
Lack of contemporary hybridization found here does not mean that admixture will not be an issue for polar bears in the future. As the Arctic warms, the species may come into contact more frequently than did the bears represented in the current study. Unchecked hybridization can become a conservation threat if the parental species collapse into a “hybrid swarm” (Levin 2002; Todesco et al. 2016). Moreover, most current legal frameworks that address species conservation are not applicable to hybrids (Allendorf et al. 2001; Fitzpatrick et al. 2015; Lind-Riehl et al. 2016). One exception is CITES (Convention on International Trade of Endangered Species), under which polar bear hybrids are treated as polar bears (Appendix II) for purposes of trade (Conf.10.17 Rev.CoP14). Thus, while widespread hybridization between polar bears and grizzly bears was not observed in this work, continued monitoring is warranted, and the genomic resources developed here can be key in these efforts. For example, the development non-invasive monitoring tools such as eDNA collected from polar bear tracks (e.g., Hellström et al. 2023) that include the species-discriminating loci on this SNP chip could increase the scale at which monitoring takes place.
The Ursus maritimus V2 SNP chip will provide a powerful tool for monitoring the adaptive potential of this species along with assessing population structure, quantitative genomics, and continued monitoring of hybridization in polar bears. This tool can provide a standardized set of loci that can be applied to all circumpolar populations allowing for easier data integration and comparisons. Such assessments may be especially critical as the polar bear is considered a species of Special Concern under the Canadian Species at Risk Act (SARA; COSEWIC 2018), which requires the completion of a species management plan. Together with other conservation initiatives (Vongraven et al. 2012, 2018; Polar Bear Range States 2015; Hamilton and Derocher 2019), the genomic resources we have developed can help inform evolutionarily enlightened management for polar bears, thereby best positioning the species to persist in the face of changing environmental conditions.
Data availability
Data is provided within the manuscript or supplementary information files.
Code availability
Not applicable.
References
Allendorf FW, Leary RF, Spruell P, Wenburg JK (2001) The problems with hybrids: setting conservation guidelines. Trends Ecol Evol 16:613–622. https://doi.org/10.1016/S0169-5347(01)02290-X
Anderson EC, Thompson EA (2002) A model-based method for identifying species hybrids using multilocus genetic data. Genetics 160:1217–1229
Andrews KR, Good JM, Miller MR et al (2016) Harnessing the power of RADseq for ecological and evolutionary genomics. Nat Rev Genet 17:81–92
Ashley MV, Willson MF, Pergams ORW et al (2003) Evolutionarily enlightened management. Biol Conserv 111:115–123. https://doi.org/10.1016/S0006-3207(02)00279-3
Bidon T, Janke A, Fain SR et al (2014) Brown and polar bear Y chromosomes reveal extensive male-biased gene flow within brother lineages. Mol Biol Evol 31:1353–1363. https://doi.org/10.1093/molbev/msu109
Cahill JA, Green RE, Fulton TL et al (2013) Genomic evidence for Island Population Conversion resolves conflicting theories of Polar Bear Evolution. PLoS Genet 9:e1003345. https://doi.org/10.1371/journal.pgen.1003345
Cahill JA, Stirling I, Kistler L et al (2015) Genomic evidence of geographically widespread effect of gene flow from polar bears into brown bears. Mol Ecol 24:1205–1217. https://doi.org/10.1111/mec.13038
Cahill JA, Heintzman PD, Harris K et al (2018) Genomic evidence of globally widespread admixture from polar bears into brown bears during the last ice age. Mol Biol Evol 35:1120–1129
Campbell E, Brunet BMT, Dupuis JR, Sperling FAH (2018) Would an RRS by any other name sound as RAD? Methods Ecol Evol 9:1920–1927
Chang C, Chow C, Tellier L et al (2015) Second-generation PLINK: rising to the challenge of larger and richer datasets. Gigascience 4: s13742-015-0047–8https://doi.org/10.1186/s13742-015-0047-8.
Clark DA, Brook R, Oliphant-Reskanski C et al (2018) Novel range overlap of three ursids in the Canadian subarctic. Arct Sci 5:62–70. https://doi.org/10.1139/as-2018-0013
Cook CN, Sgrò CM (2019) Conservation practitioners’ understanding of how to manage evolutionary processes. Conserv Biol 33:993–1001. https://doi.org/10.1111/cobi.13306
R Core Team (2019) R: A language and environment for statistical computing, reference index version 3.2.2
COSEWIC (2018) COSEWIC assessment and status report on the Polar Bear Ursus maritimus in Canada. Ottawa
Cronin MA, Rincon G, Meredith RW et al (2014) Molecular phylogeny and SNP variation of polar bears (Ursus maritimus), brown bears (U. Arctos), and black bears (U. Americanus) derived from genome sequences. J Hered 105:312–323. https://doi.org/10.1093/jhered/est133
Davey JW, Cezard T, Fuentes-Utrilla P et al (2013) Special features of RAD sequencing data: implications for genotyping. Mol Ecol 22:3151–3164. https://doi.org/10.1111/mec.12084
Derocher AE, Lunn NJ, Stirling I (2004) Polar bears in a warming climate. Integr Comp Biol 44:163–176. https://doi.org/10.1093/icb/44.2.163
Díaz-Arce N, Rodríguez-Ezpeleta N (2019) Selecting RAD-Seq Data Analysis parameters for Population Genetics: the more the Better? Front Genet 10:533
Doupé JP, England JH, Furze M, Paetkau D (2007) Most northerly observation of a grizzly bear (Ursus arctos) in Canada: photographic and DNA evidence from Melville Island, Northwest territories. Arctic 60:271–276
Duntsch L, Tomotani BM, de Villemereuil P et al (2020) Polygenic basis for adaptive morphological variation in a threatened Aotearoa, New Zealand bird, the hihi (Notiomystis cincta). Proceedings of the Royal Society B: Biological Sciences 287:20200948. https://doi.org/10.1098/rspb.2020.0948
Ellegren H (2014) Genome sequencing and population genomics in non-model organisms. Trends Ecol Evol 29:51–63. https://doi.org/10.1016/j.tree.2013.09.008
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software structure: a simulation study. Mol Ecol 14:2611–2620
Falush D, Stephens M, Pritchard JK (2003) Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics 164:1567–1587
Fitzpatrick BM, Ryan ME, Johnson JR et al (2015) Hybridization and the species problem in conservation. Curr Zool 61:206–216. https://doi.org/10.1093/czoolo/61.1.206
Frichot E, François O (2015) LEA: an R package for landscape and ecological association studies. Methods Ecol Evol 6:925–929. https://doi.org/10.1111/2041-210X.12382
Fuentes-Pardo AP, Ruzzante DE (2017) Whole-genome sequencing approaches for conservation biology: advantages, limitations, and practical recommendations. Mol Ecol 26:5369–5406. https://doi.org/10.1111/mec.14264
Funk WC, McKay JK, Hohenlohe PA, Allendorf FW (2012) Harnessing genomics for delineating conservation units. TREE 27:489–496. https://doi.org/10.1016/j.tree.2012.05.012
Gautier M, Gharbi K, Cezard T et al (2013) The effect of RAD allele dropout on the estimation of genetic variation within and between populations. Mol Ecol 22:3165–3178. https://doi.org/10.1111/mec.12089
Genetic Resources Development Consortium, Bensch S, Coltman DW et al (2014) Genomic resources notes accepted 1 June 2013-31 July 2013. Mol Ecol Resour 14:218. https://doi.org/10.1111/1755-09s98.12166
Gilbert KJ, Andrew RL, Bock DG et al (2012) Recommendations for utilizing and reporting population genetic analyses: the reproducibility of genetic clustering using the program structure. Mol Ecol 21:4925–4930. https://doi.org/10.1111/j.1365-294X.2012.05754.x
Glenn TC (2011) Field guide to next-generation DNA sequencers. Mol Ecol Resour 11:759–769
Hagen IJ, Billing AM, Rønning B et al (2013) The easy road to genome-wide medium density SNP screening in a non-model species: development and application of a 10 K SNP-chip for the house sparrow (Passer domesticus). Mol Ecol Resour 13:429–439. https://doi.org/10.1111/1755-0998.12088
Hailer F (2015) Introgressive hybridization: brown bears as vectors for polar bear alleles. Mol Ecol 24:1161–1163. https://doi.org/10.1111/mec.13101
Hamilton SG, Derocher AE (2019) Assessment of global polar bear abundance and vulnerability. Anim Conserv 22:83–95. https://doi.org/10.1111/acv.12439
Hamilton SG, de la Castro L, Derocher AE et al (2014) Projected polar bear sea ice habitat in the Canadian Arctic Archipelago. PLoS ONE 9:e113746
Harmoinen J, von Thaden A, Aspi J et al (2021) Reliable wolf-dog hybrid detection in Europe using a reduced SNP panel developed for non-invasively collected samples. BMC Genomics 22:473. https://doi.org/10.1186/s12864-021-07761-5
Harrisson KA, Pavlova A, Telonis-Scott M, Sunnucks P (2014) Using genomics to characterize evolutionary potential for conservation of wild populations. Evol Appl 7:1008–1025. https://doi.org/10.1111/eva.12149
Hassanin A (2015) The role of Pleistocene glaciations in shaping the evolution of polar and brown bears. CR Biol 338:494–501. https://doi.org/10.1016/j.crvi.2015.04.008. Evidence from a critical review of mitochondrial and nuclear genome analyses
Hellström, M, Kruger A, Näslund et al (2023) Capturing environmental DNA in snow tracks of polar bear, Eurasian lynx and snow leopard towards individual identification. Front Conserv Sci 4:1250996. https://doi.org/10.3389/fcosc.2023.1250996
Helyar SJ, Hemmer-Hansen J, Bekkevold D et al (2011) Application of SNPs for population genetics of nonmodel organisms: new opportunities and challenges. Mol Ecol Resour 11:123–136. https://doi.org/10.1111/j.1755-0998.2010.02943.x
Humble E, Paijmans AJ, Forcada J, Hoffman JI (2020) An 85K SNP array uncovers inbreeding and cryptic relatedness in an antarctic fur seal breeding colony. G3 Genes|Genomes|Genetics 10:2787–2799. https://doi.org/10.1534/g3.120.401268
Illumina (2009) Infinium® HD Assay: Ultra Protocol Guide
Janes JK, Miller JM, Dupuis JR et al (2017) The K = 2 conundrum. Mol Ecol 26:3594–3602. https://doi.org/10.1111/mec.14187
Jones MR, Good JM (2016) Targeted capture in evolutionary and ecological genomics. Mol Ecol 25:185–202. https://doi.org/10.1111/mec.13304
Judkins ME, Couger BM, Warren WC, Van Den Bussche RA (2020) A 50K SNP array reveals genetic structure for bald eagles (Haliaeetus leucocephalus). Conserv Genet 21:65–76. https://doi.org/10.1007/s10592-019-01216-x
Karlsson S, Moen T, Lien S et al (2011) Generic genetic differences between farmed and wild Atlantic salmon identified from a 7K SNP-chip. Mol Ecol Resour 11:247–253. https://doi.org/10.1111/j.1755-0998.2010.02959.x
Kawakami T, Backström N, Burri R et al (2014) Estimation of linkage disequilibrium and interspecific gene flow in Ficedula flycatchers by a newly developed 50k SNP array. Mol Ecol Resour 14:1248–1260. https://doi.org/10.1111/1755-0998.12270
Kijas JW, Porto-Neto L, Dominik S et al (2014) Linkage disequilibrium over short physical distances measured in sheep using a high-density SNP chip. Anim Genet 45:754–757. https://doi.org/10.1111/age.12197
Kopelman NM, Mayzel J, Jakobsson M et al (2015) Clumpak: a program for identifying clustering modes and packaging population structure inferences across K. Mol Ecol Resour 15:1179–1191. https://doi.org/10.1111/1755-0998.12387
Laidre KL, Born EW, Atkinson SN et al (2018) Range contraction and increasing isolation of a polar bear subpopulation in an era of sea-ice loss. Ecol Evol 8:2062–2075. https://doi.org/10.1002/ece3.3809
Laidre KL, Atkinson S, Regehr EV, Stern HL, Born EW, Wiig Ø, Lunn NJ, Dyck M (2020) Interrelated ecological impacts of climate change on an apex predator. Ecol Appl 30:4. https://doi.org/10.1002/eap.2071
Laidre KL, Supple MA, Born EW, Regehr EV, Wiig Ø, Ugarte F, Aars J, Dietz R, Sonne C, Hegelund P, Isaksen C, Akse GB, Cohen B, Stern HL, Moon T, Vollmers C, Corbett-Detig R, Paetkau D, Shapiro B (2022) Glacial ice supports a distinct and undocumented polar bear subpopulation persisting in late 21st-century sea-ice conditions. Science 376:1333–1338. https://doi.org/10.1126/science.abk2793
Lee KD, Millar CD, Brekke P et al (2021) The design and application of a 50 K SNP chip for a threatened Aotearoa New Zealand passerine, the hihi. Mol Ecol Resour 22:415–429. https://doi.org/10.1111/1755-0998.13480
Levin DA (2002) Hybridization and extinction - in protecting rare species, conservationists should consider the dangers of interbreeding, which compound the more well-known threats to wildlife. Am Sci 90:254–261. https://doi.org/10.1511/2002.9.782
Lind-Riehl JF, Mayer AL, Wellstead AM, Gailing O (2016) Hybridization, agency discretion, and implementation of the U.S. Endangered species Act. Conserv Biol 30:1288–1296. https://doi.org/10.1111/cobi.12747
Lischer HEL, Excoffier L (2011) PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 28:298–299. https://doi.org/10.1093/bioinformatics/btr642
Liu S, Lorenzen ED, Fumagalli M et al (2014) Population genomics reveal recent speciation and rapid evolutionary adaptation in polar bears. Cell 157:785–794. https://doi.org/10.1016/j.cell.2014.03.054
MacDonald ZG, Dupuis JR, Davis CS et al (2020) Gene flow and climate-associated genetic variation in a vagile habitat specialist. Mol Ecol 29:3889–3906. https://doi.org/10.1111/mec.15604
Malenfant RM (2016) Population genomics and quantitative genetics of polar bears (Ursus maritimus). PhD Thesis, University of Alberta
Malenfant RM, Coltman DW, Davis CS (2015) Design of a 9K SNP chip for polar bears (Ursus maritimus) from RAD and transcriptome sequencing. Mol Ecol Resour 15:587–600. https://doi.org/10.1111/1755-0998.12327
Malenfant RM, Davis CS, Richardson ES et al (2018) Heritability of body size in the polar bears of Western Hudson Bay. Mol Ecol Resour 18:854–866
McCue ME, Bannasch DL, Petersen JL et al (2012) A high density SNP array for the domestic horse and extant Perissodactyla: utility for association mapping, genetic diversity, and phylogeny studies. PLoS Genet 8:e1002451. https://doi.org/10.1371/journal.pgen.1002451
McMahon BJ, Teeling EC, Höglund J (2014) How and why should we implement genomics into conservation? Evol Appl 7:999–1007. https://doi.org/10.1111/eva.12193
Meek MH, Larson WA (2019) The future is now: Amplicon sequencing and sequence capture usher in the conservation genomics era. Mol Ecol Resour 19:795–803. https://doi.org/10.1111/1755-0998.12998
Miller JM, Kijas JW, Heaton MP et al (2012a) Consistent divergence times and allele sharing measured from cross-species application of SNP chips developed for three domestic species. Mol Ecol Resour 12:1145–1150. https://doi.org/10.1111/1755-0998.12017
Miller W, Schuster SC, Welch AJ et al (2012b) Polar and brown bear genomes reveal ancient admixture and demographic footprints of past climate change. Proceedings of the National Academy of Sciences 109:E2382–E2390. https://doi.org/10.1073/pnas.1210506109
Molnár PK, Bitz CM, Holland MM et al (2020) Fasting season length sets temporal limits for global polar bear persistence. Nat Clim Change 10:732–738. https://doi.org/10.1038/s41558-020-0818-9
Morin PA, Luikart G, Wayne RK, Group SNPW (2004) SNPs in ecology, evolution and conservation. Trends Ecol Evol 19:208–216
Nielsen EE, Bach LA, Kotlicki P (2006) HYBRIDLAB (version 1.0): a program for generating simulated hybrids from population samples. Mol Ecol Notes 6:971–973. https://doi.org/10.1111/j.1471-8286.2006.01433.x
Oliphant A, Barker DL, Stuelpnagel JR, Chee MS (2002) BeadArray technology: enabling an accurate, cost-effective approach to high-throughput genotyping. Biotechniques 32
Peterson BK, Weber JN, Kay EH et al (2012) Double digest RADseq: an inexpensive method for de novo snp discovery and genotyping in model and non-model species. PLoS ONE 7:e37135
Polar Bear Range States (2015) Circumpolar action plan: conservation strategy for polar bears. A product of the representatives of the parties to the 1973. Agreement on the Conservation of Polar Bears
Pongracz JD, Paetkau D, Branigan M, Richardson E (2017) Recent hybridization between a polar bear and grizzly bears in the Canadian arctic. Arctic 70:151–160. https://doi.org/10.14430/arctic4643
Pritchard JK, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Purcell S, Neale B, Todd-Brown K et al (2007) PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 81:559–575
Rockwell R, Gormezano L, Hedman D (2008) Grizzly bears, Ursus arctos, in Wapusk National Park, Northeastern Manitoba. Can Field-Naturalist 122:323–326
Sgrò CM, Lowe AJ, Hoffmann AA (2011) Building evolutionary resilience for conserving biodiversity under climate change. Evol Appl 4:326–337. https://doi.org/10.1111/j.1752-4571.2010.00157.x
Shafer ABA, Peart CR, Tusso S et al (2017) Bioinformatic processing of RAD-seq data dramatically impacts downstream population genetic inference. Methods Ecol Evol 8:907–917. https://doi.org/10.1111/2041-210X.12700
Shen R, Fan J-B, Campbell D et al (2005) High-throughput SNP genotyping on universal bead arrays. Mutat Research/Fundamental Mol Mech Mutagen 573:70–82
Song Q, Hyten DL, Jia G et al (2013) Development and evaluation of SoySNP50K, a high-density genotyping array for soybean. PLoS ONE 8:e54985. https://doi.org/10.1371/journal.pone.0054985
Stirling I, Derocher AE (2012) Effects of climate warming on polar bears: a review of the evidence. Glob Change Biol 18:2694–2706. https://doi.org/10.1111/j.1365-2486.2012.02753.x
Suchan T, Espíndola A, Rutschmann S et al (2017) Assessing the potential of RAD-sequencing to resolve phylogenetic relationships within species radiations: the fly genus Chiastocheta (Diptera: Anthomyiidae) as a case study. Mol Phylogenet Evol 114:189–198. https://doi.org/10.1016/j.ympev.2017.06.012
Todesco M, Pascual MA, Owens GL et al (2016) Hybridization and extinction. Evol Appl 9:892–908. https://doi.org/10.1111/eva.12367
van Dijk EL, Jaszczyszyn Y, Naquin D, Thermes C (2018) The third revolution in sequencing technology. Trends Genet 34:666–681. https://doi.org/10.1016/j.tig.2018.05.008
Vongraven D, Aars J, Amstrup S et al (2012) A circumpolar monitoring framework for polar bears. Ursus 23:1–66. https://doi.org/10.2192/URSUS-D-11-00026.1
Vongraven D, Derocher AE, Bohart A (2018) Polar bear research: has science helped management and conservation? Environ Reviews 26:358–368. https://doi.org/10.1139/er-2018-0021
Wang M-S, Murray GGR, Mann D et al (2022) A polar bear paleogenome reveals extensive ancient gene flow from polar bears into brown bears. Nat Ecol Evol. https://doi.org/10.1038/s41559-022-01753-8
Wringe BF, Stanley RRE, Jeffery NW et al (2017) Parallelnewhybrid: an r package for the parallelization of hybrid detection using newhybrids. Mol Ecol Resour 17:91–95. https://doi.org/10.1111/1755-0998.12597
Acknowledgements
Any use of trade, firm, or product names is for descriptive purposes only and does not imply endorsement by the U.S. Government. Thanks go to Colin Coros, Dr. David Coltman, and Dr. Janice Cooke for providing computing infrastructure used in the analyses and to Dr. David Paetkau for providing DNA extractions for historical samples to be included in this work. We would like to thank all the Inuit hunters across the circumpolar Arctic that have contributed to this study as well as past studies to further our understanding of the evolutionary ecology of polar bears. Thanks also go to the hunters, indigenous organizations, and government monitoring programs that supported data collections in Northwest Territories and Nunavut.
Funding
Open access funding provided by Environment & Climate Change Canada library. Funding from Environment and Climate Change Canada (ECCC) provided by the Wildlife Research Division. Illumina BeadChip development and genotyping supported by the Strategic Technology Applications of Genomics in the Environment (STAGE) funding program. Funding from the U.S. Geological Survey was provided through the Ecosystems Mission Area and the Changing Arctic Ecosystems Initiative. Funding for the polar bear sampling in East Greenland in 2007 and 2008 conducted by Greenland Institute of Natural Resources, University of Oslo, and Aarhus University were provided by the Bureau of Mineral and Petroleum (Nuuk, Greenland), the Danish Ministry of the Environment (Copenhagen, Denmark), and the Greenland Institute of Natural Resources (Nuuk).
Open access funding provided by Environment & Climate Change Canada library.
Author information
Authors and Affiliations
Contributions
Conceptualization: JMM Data curation: JMM, RMM, KLL, JP, ESR, CSD. Formal analysis and investigation: JMM, LRR. Writing - original draft preparation: JMM. Writing - review and editing: RMM, LRR, KLL, EWB, RD, SB, JP, ESR, TCA, ØW, CSD. Funding acquisition: ESR, CSD. Resources: KLL, EWB, RD, JP, SB, ESR, ØW, CSD.
Corresponding author
Ethics declarations
Ethical approval
Environment and Climate Change Canada (ECCC) sample collections were reviewed and approved by the ECCC Western and Northern Animal Care Committee. Northwest Territories sample collections were reviewed and approved by the Wildlife Care Committee, Northwest Territories. U.S. Geological Survey polar bear research was conducted under U.S. Fish and Wildlife Service permit #MA690038, and capture and handling protocols were approved by the U.S. Geological Survey Alaska Science Center Institutional Animal Care and Use Committee. Samples from Greenland were collected under approvals J.nr. 66.24/03 (5 February 2007 and 5 February 2008) and 2015-110281/document 1858991 from the Greenland Ministry for Hunting, Fisheries and Agriculture. Additional samples were collected as part of subsistence harvest by Inuit and therefore exempt from institutional ethical approval.
Consent to participate
Not applicable.
Consent for publication
All authors have read the manuscript and approve of its submission for publication.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Miller, J.M., Malenfant, R.M., Rivkin, L.R. et al. Development of an 8K SNP chip to assess adaptive diversity and hybridization in polar bears. Conservation Genet Resour (2024). https://doi.org/10.1007/s12686-024-01359-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12686-024-01359-1