Abstract
Metabarcoding is emerging as an alternative to morphological methods in noninvasive carnivore diet analysis based on scats. A number of metabarcoding markers have been developed but their comparative performance to recover DNA from scats remains mostly untested. We tested three markers covering a wide taxonomic range of prey items and compared them with the results of a morphological analysis. Morphological and genetic methods performed comparably regarding the identity of detected prey species, but the number of identified species varied strongly between markers. Only one, 12S-V5, amplified successfully in all samples and proved to be robust and reliable when working with the highly degraded DNA obtained from scats.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Main text
Carnivore diet composition is often inferred by morphological analysis of prey remains in scats, but metabarcoding as a genetic alternative is used with increasing frequency (Monterroso et al. 2019; Shi et al. 2021). For animals in general, the standard metabarcoding marker is a region of the COI gene while specialized vertebrate markers target mitochondrial 12S ribosomal RNA. Currently available markers were developed to meet the requirements of different DNA sources but their performance regarding mammalian prey species identification based on scats has usually not been assessed (Ando et al. 2020; Kocher et al. 2017).
To address this issue, we compared the performance of three different metabarcoding markers (Table 1) in wolf prey item identification from scat samples and compared them with the results obtained by morphological prey item analysis. The very short 12S-V5 (12S, ca. 100 bp, Riaz et al. 2011) was specifically developed for use with highly degraded fecal samples. In comparison, the longer Mam12S-340 (12S, ca. 340 bp, Kocher et al. 2017) and mlCOIint-F/jgHCO2198-R (CO1, ca. 310 bp, Leray et al. 2013; Geller et al. 2013) offer higher resolution to identify closely related vertebrate species (Mam12S-340) and a wider phylogenetic range, covering all animals as well as fungi and bacteria (mlCOIint-F/jgHCO2198-R) but have not been tested with fecal samples so far.
We started with 24 wolf fecal samples collected in the Glücksburger Heide, Saxonia-Anhalt, Germany. Detailed descriptions of study site, collection method, and morphological scat analysis are given in Lippitsch et al. (2024). Prior to morphological analysis, DNA samples were taken from both ends of each scat. DNA was extracted using the QIAamp Fast DNA Stool Mini Kit (QIAGEN, Hilden, Germany).
For all samples, we tested successful PCR amplification using agarose gel electrophoresis, eight samples that showed successful amplification were selected for metabarcoding. Wolf-specific blocking primers do not increase the number of prey items detected (Shi et al. 2021) and were therefore not used.
Sequencing was done on the Illumina MiSeq platform by LGC (Berlin, Germany). All samples and markers were sequenced individually. Read processing and amplicon filtering were done using Vsearch (Rognes et al. 2016). We used the following filtering commands and parameters: quality filtering (--fastq_maxee 1), denoising (--minsize 4 --unoise_alpha 2), indel filtering (--fastq_minlen [10% smaller than anticipated fragment length] --fastq_maxlen [10% larger than anticipated fragment length]), chimera filtering (--uchime3_denovo --nonchimeras). To account for possible contamination, sequences with a read count below 0.05% of the total read count of the respective sample were suppressed. Taxonomic assignment was done by using blastn (Johnson et al. 2008) against the NCBI GenBank database, applying a 0.99% of identity threshold.
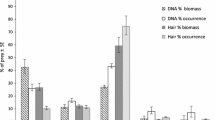
Prey items identified within each sample are shown in Table 2. Despite successful amplification of wolf DNA, five samples failed to retrieve any prey items for Mam12S-340, and two for COI. For 12S-V5, prey item detection succeeded in all samples.
While the identity of detected prey species was comparable between markers, the number of identified prey items and reads per item varied strongly (Table 2). Morphological and genetic results were comparable and corresponded well with those from an exhaustive dietary study in the study area (Lippitsch et al. 2024). There were, however, some notable differences. In sample 5, the metabarcoding was able to identify the species of an item that could only be identified to family level morphologically. In samples 6 and 7, morphological and genetic results seem to differ. The morphological analysis identified only Capreolus capreolus while all metabarcodes unambiguously identified Cervus elaphus as the main prey item. In both cases, however, the genetic analysis picked up traces of C. capreolus. As material for the genetic analysis was taken from the ends of the scat, it is plausible that the metabarcoding detected the content of a different meal than the morphological analysis which studied the entire scat. The occurrence of fox (Vulpes vulpes) in two samples can likely be explained by the fact that foxes mark wolf scat, thereby contaminating the sample. While foxes occasionally form part of wolf diet (Nowak et al. 2011; Wagner et al. 2012), no macroscopic remains were identified in the respective samples. The identification of a vole in sample 6 reinforces the sensitivity of the marker.
Our study demonstrates differences in the performance of these three markers that would strongly affect the success of studies aimed at identifying prey items based on highly degraded DNA from scats. Very short metabarcodes like 12S-V5 seem to be the only viable option for this difficult material. The longer Mam12S-340 and COI metabarcodes were developed for the analysis of gut content which, unlike scats, was not subjected to environmental effects. If the broader phylogenetic range of the COI region is needed, Kocher et al. (2017) designed a short COI-metabarcode. To identify plant food items, which formed an important component in sample 6, a short metabarcode based on the TrnL-region developed by Taberlet et al. (2007) is promising (Boukhdoud et al. 2021). For studies focusing on vertebrates, 12S-V5 has proven to be a robust and sensitive marker. It offers an alternative to the morphological analysis of diet composition in carnivores and can expand the range of identifiable prey species in regard to smaller species.
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Ando H, Mukai H, Komura T, Dewi T, Ando M, Isagi Y (2020) Methodological trends and perspectives of animal dietary studies by noninvasive fecal DNA metabarcoding. Environ DNA 2:391–406. https://doi.org/10.1002/edn3.117
Boukhdoud L, Saliba C, Parker LD, McInerney NR, Kahale R, Saliba I, Maldonado JE, Kharrat MBD (2021) Using DNA metabarcoding to decipher the diet plant component of mammals from the Eastern Mediterranean region. Metabarcoding Metagenom 5:e70107. https://doi.org/10.3897/mbmg.5.70107
Geller J, Meyer C, Parker M, Hawk H (2013) Redesign of PCR primers for mitochondrial cytochrome c oxidase subunit I for marine invertebrates and application in all-taxa biotic surveys. Mol Ecol Resour 13:851–861. https://doi.org/10.1111/1755-0998.12138
Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL (2008) NCBI BLAST: a better web interface. Nucleic Acids Res 36:W5–W9. https://doi.org/10.1093/nar/gkn20
Kocher A, de Thoisy B, Catzeflis F, Huguin M, Valière S, Zinger L, Bañuls A-L, Murienne J (2017) Evaluation of short mitochondrial metabarcodes for the identification of amazonian mammals. Methods Ecol Evol 8:1276–1283. https://doi.org/10.1111/2041-210X.12729
Leray M, Yang JY, Meyer CP, Mills SC, Agudelo N, Ranwez V, Boehm JT, Machida RJ (2013) A new versatile primer set targeting a short fragment of the mitochondrial COI region for metabarcoding metazoan diversity: application for characterizing coral reef fish gut contents. Front Zool 10:34. https://doi.org/10.1186/1742-9994-10-34
Lippitsch P, Kühl H, Reinhardt I, Kluth G, Böcker F, Kruk M, Michler FU, Schumann H, Teubner Ja, Teubner Je, Trost M, Weber H, Ansorge A (2024) Feeding dynamics of the wolf (Canis lupus) in the anthropogenic landscape of Germany: a 20-year survey. Mamm Biol 104:151–163. https://doi.org/10.1007/s42991-024-00399-2
Monterroso P, Godinho R, Oliveira T, Ferreras P, Kelly MJ, Morin DJ, Waits LP, Alves PC, Mills LS (2019) Feeding ecological knowledge: the underutilised power of faecal DNA approaches for Carnivore diet analysis. Mam Rev 49:97–112. https://doi.org/10.1111/mam.12144
Nowak S, Mysłajek RW, Kłosińska A, Gabryś G (2011) Diet and prey selection of wolves (Canis lupus) recolonising western and Central Poland. Mamm Biol 76:709–715. https://doi.org/10.1016/j.mambio.2011.06.007
Riaz T, Shehzad W, Viari A, Pompanon F, Taberlet P, Coissac E (2011) ecoPrimers: inference of new DNA barcode markers from whole genome sequence analysis. Nucleic Acids Res 39:e145. https://doi.org/10.1093/nar/gkr732
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. https://doi.org/10.7717/peerj.2584
Shi Y, Hoareau Y, Reese EM et al (2021) Prey partitioning between sympatric wild carnivores revealed by DNA metabarcoding: a case study on wolf (Canis lupus) and coyote (Canis latrans) in northeastern Washington. Conserv Genet 22:293–305. https://doi.org/10.1007/s10592-021-01337-2
Taberlet P, Coissac E, Pompanon F, Gielly L, Miquel C, Valentini A, Vermat T, Corthier G, Brochmann C, Willerslev E (2007) Power and limitations of the chloroplast trnL (UAA) intron for plant DNA barcoding. Nucleic Acids Res 35:e14. https://doi.org/10.1093/nar/gkl938
Wagner C, Holzapfel M, Kluth G, Reinhardt I, Ansorge H (2012) Wolf (Canis lupus) feeding habits during the first eight years of its occurrence in Germany. Mamm Biol 77:196–203. https://doi.org/10.1016/j.mambio.2011.12.004
Acknowledgements
The authors are grateful to the volunteers who contributed to the wolf scat collecting in Saxony-Anhalt and Brandenburg, whose voluntary commitment forms an important basis for this study.
Funding
Open Access funding enabled and organized by Projekt DEAL. The authors did not receive external support from any organization for the submitted work.
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Contributions
P.E. originally conceived and designed the study. All authors contributed to the study design. Material preparation, data collection and morphological dietary analysis were performed by P.L. and H.S. Molecular lab work, data collection and genetic analysis were performed by V.K. and J.R. Analysis and interpretation of the metabarcoding results were performed by J.R. and P.E. The first draft of the manuscript was written by P.E. and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Eusemann, P., Rees, J., Kuhlenkamp, V. et al. Dietary analysis of wolf (Canis lupus) – a comparison of markers and methods. Conservation Genet Resour (2024). https://doi.org/10.1007/s12686-024-01356-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s12686-024-01356-4